Abstract
The Freiburg RNA tools web server integrates three tools for the advanced analysis of RNA in a common web-based user interface. The tools IntaRNA, ExpaRNA and LocARNA support the prediction of RNA–RNA interaction, exact RNA matching and alignment of RNA, respectively. The Freiburg RNA tools web server and the software packages of the stand-alone tools are freely accessible at http://rna.informatik.uni-freiburg.de.
INTRODUCTION
During the last decade, the discovery of a multitude of regulatory and catalytic RNA molecules has attracted attention to RNA in biological research (1,2). Many non-coding RNAs (ncRNAs) require a specific structure to perform their functions or interact via structural base pairing (3–6). Thus, RNA analysis demands tools that take into account RNA structure. The Freiburg RNA tools web server gives access to tools for three advanced RNA analysis tasks via an integrated, easy to use interface that supports the combination of these tools.
The server offers the prediction of RNA–RNA interaction (IntaRNA), exact RNA matching (ExpaRNA), and the multiple alignment of RNA (LocARNA). All tools are recently developed, continuously maintained, highly accurate and among the best of their class (7–9). Consequently, the tools have been used in recent studies (10–12).
Performing complex analysis tasks, the server complements available web servers for RNA analysis such as the Vienna RNA web suite (13) and the mfold web server (14). Among other services, the Vienna RNA web suite gives access to an older version of LocARNA. In this contribution, we offer increased functionality with improved performance.
GENERAL OVERVIEW
The central purpose of the web server is to provide RNA analysis tools that have been developed by the Freiburg Bioinformatics Group. To this end, the web server integrates three tools for different analysis tasks in a common framework.
Each tool accepts a set of sequences in FASTA format as its main input. These sequences can be either entered directly or uploaded. The tools allow specific extension of the FASTA format, such as the annotation of sequences by secondary structure in dot–bracket format. Furthermore, each input page provides program-specific options, with reasonable default settings, in order that the user can configure the respective tool to their needs. For user convenience, the server distinguishes between basic options and advanced parameters. The latter are hidden by default and can be unfolded on demand. In this way the server provides broad flexibility without confusing the less experienced user. Input is validated and the user is informed of inconsistencies as early as possible. For each task, we provide example input for demonstration purposes. Online help is provided and describes each tool, its input, available options and output.
Tool output and method are specific to each task and are, therefore, described separately. Where possible the output is illustrated by graphical presentation of the results. Figures are displayed in the browser as pngs and offered for download in postscript and pdf format.
Each analysis task is processed following a general scheme: jobs are scheduled to a computing cluster in order that jobs can be computed in parallel and resources flexibly adapted to the server load. Currently, we reserve eight cores for parallel computation. After submission, the current status of the job is reported and the user receives a URL allowing access to the job status or output. Upon job completion the result page is displayed online in the web browser.
Finally, the server provides links to the source code of the tools. The stand-alone command-line versions are more convenient and appropriate for large-scale studies; however there are no input size restrictions by our web server. All tools use the widely accepted Turner free–energy model for RNA folding with standard energy parameters (15).
IntaRNA
IntaRNA is a tool for fast and accurate prediction of interactions between two RNA molecules (7). It has been designed to predict mRNA target sites for ncRNAs like eukaryotic microRNAs or bacterial small RNAs (sRNAs), but it can also be used to predict other RNA–RNA interactions.
Input and output
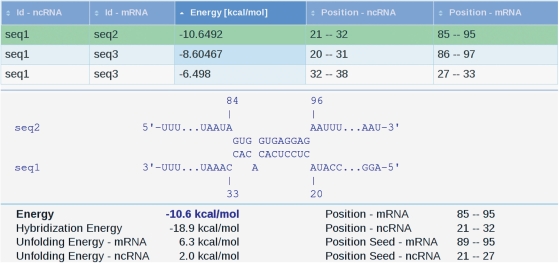
The input of IntaRNA is a set of ncRNA sequences and a set of mRNA sequences. The output consists of a table that summarizes the results of the prediction and links to all predicted putative interactions between the ncRNAs and mRNAs. The output table can be sorted by columns to allow selection of interactions by sequence identifier, interaction energy score or interaction position in each sequence (Figure 1).
Figure 1.
Screenshot of the IntaRNA result page for a set of example sequences. The table summarizes all predicted interactions including one suboptimal interaction for mRNA seq3. It can be sorted by clicking on the header of a column. The interaction shown below the table is highlighted in green. For this prediction, additional information such as interaction positions and different contributions of the interaction energy score are given.
Method
IntaRNA computes interactions by minimizing an interaction energy score via dynamic programming. The scoring is based on the hybridization free energy and accessibility of the interacting subsequences. The accessibility of an interaction site is defined as the free energy that is required to make the interaction site single-stranded. This is based on the thermodynamic ensemble of all secondary structures. Computation of these accessibilities is realized via the Vienna RNA library (16–18). It is assumed that ncRNAs fold globally and that mRNAs fold locally with a given maximal base pair distance. The algorithm runs in 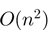 time and space when accessibilities are pre-computed.
time and space when accessibilities are pre-computed.
Furthermore, IntaRNA enables the inclusion of an interaction seed, i.e. an initial interaction region of (nearly) perfect sequence complementarity. The user has to specify the minimal number of perfectly paired bases and the maximal number of unpaired bases in the seed region. Other seed features, such as the seed position in the ncRNA, are optional.
In addition to the optimal solution according to the interaction energy score, IntaRNA optionally reports also suboptimal interactions. The user can specify the maximal number of suboptimal predictions per sequence pair or restrict the reported interactions by an energy threshold.
Prior application and evaluation
IntaRNA was validated on a data set of 18 experimentally verified sRNA–mRNA interactions, on which it achieved the highest accuracy, of all compared methods, in terms of sensitivity and positive predictive value (7). In a genome-wide target search, IntaRNA showed the best prediction performance together with the comparable approach RNAup (17), but with considerably lower computing time and memory requirement (7). Recently, IntaRNA was applied to identify two novel mRNA targets of the cyanobacterial RNA Yfr1 (10).
ExpaRNA
ExpaRNA is a tool for very fast comparison of RNAs by exact local matches (8,20). Instead of computing a full sequence-structure alignment, ExpaRNA efficiently computes the best arrangement of sequence–structure motifs common to two RNAs. This approach is beneficial for comparative sequence analysis in biology and in high-throughput RNA analysis tasks. ExpaRNA elucidates information about identical structural motifs. This is not directly addressed by sequence–structure alignment tools and, therefore, may remain hidden. In addition, the predicted set of motifs can be used as anchor constraints to speed up and guide Sankoff-style alignment methods like LocARNA and related approaches that are in principle able to profit from alignment constraints.
Input and output
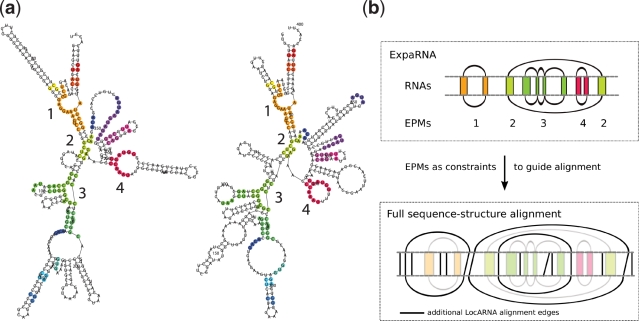
The input of ExpaRNA is a pair of RNA sequences and secondary structures in dot–bracket notation using an extended FASTA format. If no secondary structure is available, the sequences are automatically folded by RNAfold (16). ExpaRNA outputs the optimal set of exact pattern matches (EPMs) between the input RNAs. The result is presented graphically as coloured secondary structure plots (Figure 2a). Additionally, the web server allows the user to download results in different text file formats, e.g. as a structure annotated alignment or a list of (all) exact matches.
Figure 2.
(a) Annotated structures from the ExpaRNA output. Regions of exact pattern matches (EPMs) have the same colour. Shown are two bacterial RNase P RNAs (left: A-type P RNA from Escherichia coli; right: B-type P RNA from Bacillus subtilis). Structures are taken from RNase P database (19). The numbers indicate four large EPMs. (b) Workflow for combining ExpaRNA with LocARNA. First ExpaRNA is called on the input RNAs to predict EPMs 1–4. This information is used as anchor constraints for a complete sequence–structure alignment by LocARNA. EPM numbers and colours are taken from (a).
Method
ExpaRNA performs a fast pre-processing step that determines the set of all possible EPMs for two given RNAs (21). An EPM is a local substructure that is identical in sequence and structure to both RNAs. EPMs are maximally extended and bond preserving, but the set of all EPMs contains overlapping and crossing EPMs. Therefore, in the next step ExpaRNA computes the best set of non-crossing and non-overlapping EPMs, i.e. the longest collinear sequence of exact matching substructures for two RNAs. The dynamic programming algorithm runs in 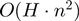 time and
time and 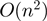 space with
space with 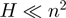 for real RNA structures.
for real RNA structures.
Prior application and evaluation
ExpaRNA results agree well with existing alignment-based methods like RNAforester (22), but results are obtained in a fraction of the compared run time. The performance of ExpaRNA combined with LocARNA was evaluated on BRAliBase 2.1 (23) and gave an overall speed-up of 4.25× with an alignment accuracy close to LocARNA alone (8) .
Combining ExpaRNA and LocARNA
ExpaRNA’s exact matches can be beneficially used as anchor constraints for a full sequence–structure alignment. This allows the calculation of a constraint alignment by LocARNA, hence enabling alignment of very large RNAs that could not otherwise be aligned in reasonable time. This approach also maintains existing structural motifs in the resulting alignment (Figure 2b). This procedure is supported by the web server with a direct link from the ExpaRNA results page to the LocARNA input page.
LocARNA
LocARNA is a tool for aligning multiple RNA sequences (9). It is one of the fastest and most accurate tools for this purpose. Comparable with programs like Dynalign, FoldAlign and Lara (24–26), LocARNA performs Sankoff-style simultaneous alignment and folding (27). Such programs generate high quality alignments that take structural similarity into account. Notably, the structural information is not required a priori but can be inferred, in parallel to the alignment process, based on an RNA free–energy model.
Input and output
The input of LocARNA is a set of sequences in FASTA format. Optionally the input can be enriched by structural constraints and anchor constraints. These constraints are useful for guiding the automatic alignment using prior knowledge and/or speeding up the computation.
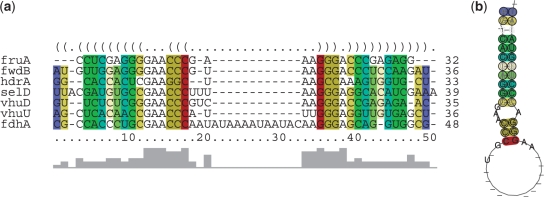
The output is a multiple alignment of these sequences optimized according to the LocARNA score, which evaluates both sequence and structural similarity (Figure 3a). The output alignment optionally satisfies given constraints. When performing local alignment, LocARNA will allow unaligned fragments at the beginning and end of all sequences without penalty. The LocARNA alignment is shown together with the predicted structure, using the alignment as input, by RNAalifold (28) (Figure 3b).
Figure 3.
Figures from the LocARNA result page of an example aligning SECIS elements. (a) Alignment with colour annotation showing the conservation of base pairs and annotated column identity as generated by RNAalifold. (b) 2D plot of consensus secondary structure predicted from (a), using the same colour scheme.
Method
LocARNA computes pairwise alignments by dynamic programming. Multiple alignments are constructed from pairwise alignments using a progressive alignment strategy. LocARNA achieves its low time and space complexity of 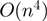 and
and 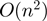 , respectively, for pairwise alignment because it needs to consider only significant base pairs (9).
, respectively, for pairwise alignment because it needs to consider only significant base pairs (9).
Prior application and evaluation
A prior LocARNA version was validated by a re-clustering of Rfam (9). At high average recall, the Rfam families were reproduced with good precision. Furthermore, LocARNA was benchmarked using BRAliBase 2.1 (23) for multiple alignment. It performed better than the comparable approaches FoldAlign and Lara (29).
IMPLEMENTATION
The Freiburg RNA tools web server is based on a general framework developed for the CPSP web tools server (30) and has been continuously improved. XHTML is served by Apache Tomcat v5.5.25 that supports the use of JavaServer Pages and Java Servlets consequently allowing a large deal of dynamically generated content to be provided.
JavaScripting is used to aid the user in providing well-formed input (sequences and parameters), which is then stored in a Java Bean and processed by a Java Servlet. Jobs with valid input parameters are submitted to a computing cluster managed by Sun Grid Engine and the user is directed to the results page to wait for job completion. Upon completion of a job, the user can access result details consisting of ‘system call’ parameters saved by the Java Bean, raw tool result data and post-processed result data (e.g. images generated from raw tool results by a Java Servlet). JavaScript, JSP and JavaServlet pages are used to increase accessibility of result features. The whole web server is run on a virtual machine hosted on a server running Scientific Linux.
FUNDING
German Research Foundation (DFG) (BA 2168/2-1 to A.S.R. and R.B., BA 2168/3-1 to R.B., BA 2168/4-1 to R.B. and WI 3628/1-1 to S.B.); German Federal Ministry of Education and Research (BMBF) (0313921 to R.B. and S.H.). Funding for open access charge: University of Freiburg.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We thank Dragoş Alexandru Sorescu, Martin Mann and Stefan Jankowski for their assistance in setting up the web server. We also thank all people who have been involved in testing the robustness and functionality of the final web server.
REFERENCES
- 1.Sharp PA. The centrality of RNA. Cell. 2009;136:577–580. doi: 10.1016/j.cell.2009.02.007. [DOI] [PubMed] [Google Scholar]
- 2.Amaral PP, Dinger ME, Mercer TR, Mattick JS. The eukaryotic genome as an RNA machine. Science. 2008;319:1787–1789. doi: 10.1126/science.1155472. [DOI] [PubMed] [Google Scholar]
- 3.Washietl S, Pedersen JS, Korbel JO, Stocsits C, Gruber AR, Hackermüller J, Hertel J, Lindemeyer M, Reiche K, Tanzer A, et al. Structured RNAs in the ENCODE selected regions of the human genome. Genome Res. 2007;17:852–864. doi: 10.1101/gr.5650707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Serganov A, Patel DJ. Ribozymes, riboswitches and beyond: regulation of gene expression without proteins. Nat. Rev. Genet. 2007;8:776–790. doi: 10.1038/nrg2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mattick JS, Makunin IV. Non-coding RNA. Hum. Mol. Genet. 2006;15:R17–R29. doi: 10.1093/hmg/ddl046. [DOI] [PubMed] [Google Scholar]
- 6.Fröhlich KS, Vogel J. Activation of gene expression by small RNA. Curr. Opin. Microbiol. 2009;12:674–682. doi: 10.1016/j.mib.2009.09.009. [DOI] [PubMed] [Google Scholar]
- 7.Busch A, Richter AS, Backofen R. IntaRNA: efficient prediction of bacterial sRNA targets incorporating target site accessibility and seed regions. Bioinformatics. 2008;24:2849–2856. doi: 10.1093/bioinformatics/btn544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Heyne S, Will S, Beckstette M, Backofen R. Lightweight comparison of RNAs based on exact sequence-structure matches. Bioinformatics. 2009;25:2095–2102. doi: 10.1093/bioinformatics/btp065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Will S, Reiche K, Hofacker IL, Stadler PF, Backofen R. Inferring non-coding RNA families and classes by means of genome-scale structure-based clustering. PLoS Comput. Biol. 2007;3:e65. doi: 10.1371/journal.pcbi.0030065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Richter AS, Schleberger C, Backofen R, Steglich C. Seed-based IntaRNA prediction combined with GFP-reporter system identifies mRNA targets of the small RNA Yfr1. Bioinformatics. 2010;26:1–5. doi: 10.1093/bioinformatics/btp609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gruber AR, Kilgus C, Mosig A, Hofacker IL, Hennig W, Stadler PF. Arthropod 7SK RNA. Mol. Biol. Evol. 2008;25:1923–1930. doi: 10.1093/molbev/msn140. [DOI] [PubMed] [Google Scholar]
- 12.Rose D, Hackermüller J, Washietl S, Reiche K, Hertel J, Findeiss S, Stadler PF, Prohaska SJ. Computational RNomics of drosophilids. BMC Genomics. 2007;8:406. doi: 10.1186/1471-2164-8-406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gruber AR, Lorenz R, Bernhart SH, Neubock R, Hofacker IL. The Vienna RNA websuite. Nucleic Acids Research. 2008;36:W70–W74. doi: 10.1093/nar/gkn188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mathews D, Sabina J, Zuker M, Turner D. Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J. Mol. Biol. 1999;288:911–940. doi: 10.1006/jmbi.1999.2700. [DOI] [PubMed] [Google Scholar]
- 16.Hofacker IL, Fontana W, Stadler PF, Bonhoeffer S, Tacker M, Schuster P. Fast folding and comparison of RNA secondary structures. Monatsh. Chem. 1994;125:167–188. [Google Scholar]
- 17.Mückstein U, Tafer H, Bernhart SH, Hernandez-Rosales M, Vogel J, Stadler PF, Hofacker IL. Translational control by RNA-RNA interaction: improved computation of RNA-RNA binding thermodynamics. In: Elloumi M, Küng J, Linial M, Murphy R, Schneider K, Toma C, editors. Bioinformatics Research and Development, of Communications in Computer and Information Science. Vol. 13. Heidelberg, Berlin, Germany: Springer-Verlag; 2008. pp. 114–127. [Google Scholar]
- 18.Bernhart SH, Hofacker IL, Stadler PF. Local RNA base pairing probabilities in large sequences. Bioinformatics. 2006;22:614–615. doi: 10.1093/bioinformatics/btk014. [DOI] [PubMed] [Google Scholar]
- 19.Brown JW. The ribonuclease P database. Nucleic Acids Res. 1999;27:314. doi: 10.1093/nar/27.1.314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Heyne S, Will S, Beckstette M, Backofen R. Proceedings of the German Conference on Bioinformatics (GCB'2008) 2008. Vol. P-136 of Lecture Notes in Informatics (LNI), Gesellschaft für Informatik (GI), Bonn, Germany, pp. 189–198. [Google Scholar]
- 21.Backofen R, Siebert S. Fast detection of common sequence structure patterns in RNAs. J. Discrete Algorithms. 2007;5:212–228. [Google Scholar]
- 22.Höchsmann M, Töller T, Giegerich R, Kurtz S. Proceedings of Computational Systems Bioinformatics (CSB 2003) 2003. Vol. 2, IEEE Computer Society, Los Alamitos, CA, USA, pp. 159–168. [Google Scholar]
- 23.Wilm A, Mainz I, Steger G. An enhanced RNA alignment benchmark for sequence alignment programs. Algorithms Mol. Biol. 2006;1:19. doi: 10.1186/1748-7188-1-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Harmanci AO, Sharma G, Mathews DH. Efficient pairwise RNA structure prediction using probabilistic alignment constraints in Dynalign. BMC Bioinformatics. 2007;8:130. doi: 10.1186/1471-2105-8-130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Torarinsson E, Havgaard JH, Gorodkin J. Multiple structural alignment and clustering of RNA sequences. Bioinformatics. 2007;23:926–932. doi: 10.1093/bioinformatics/btm049. [DOI] [PubMed] [Google Scholar]
- 26.Bauer M, Klau GW, Reinert K. Accurate multiple sequence-structure alignment of RNA sequences using combinatorial optimization. BMC Bioinformatics. 2007;8:271. doi: 10.1186/1471-2105-8-271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sankoff D. Simultaneous solution of the RNA folding, alignment and protosequence problems. SIAM J. Appl. Math. 1985;45:810–825. [Google Scholar]
- 28.Bernhart SH, Hofacker IL, Will S, Gruber AR, Stadler PF. RNAalifold: improved consensus structure prediction for RNA alignments. BMC Bioinformatics. 2008;9:474. doi: 10.1186/1471-2105-9-474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Otto W, Will S, Backofen R. Proceedings of German Conference on Bioinformatics (GCB'2008) 2008. Vol. P-136 of Lecture Notes in Informatics (LNI), Gesellschaft für Informatik (GI), Bonn, Germany, pp. 178–188. [Google Scholar]
- 30.Mann M, Smith C, Rabbath M, Edwards M, Will S, Backofen R. CPSP-web-tools: a server for 3D lattice protein studies. Bioinformatics. 2009;25:676–677. doi: 10.1093/bioinformatics/btp034. [DOI] [PMC free article] [PubMed] [Google Scholar]