Abstract
Frog is a web tool dedicated to small compound 3D generation. Here we present the new version, Frog2, which allows the generation of conformation ensembles of small molecules starting from either 1D, 2D or 3D description of the compounds. From a compound description in one of the SMILES, SDF or mol2 formats, the server will return an ensemble of diverse conformers generated using a two stage Monte Carlo approach in the dihedral space. When starting from 1D or 2D description of compounds, Frog2 is capable to detect the sites of ambiguous stereoisomery, and thus to sample different stereoisomers. Frog2 also embeds new energy minimization and ring generation facilities that solve the problem of some missing cycle structures in the Frog1 ring library. Finally, the optimized generator of conformation ensembles in Frog2 results in a gain of computational time permitting Frog2 to be up to 20 times faster that Frog1, while producing satisfactory conformations in terms of structural quality and conformational diversity. The high speed and the good quality of generated conformational ensembles makes it possible the treatment of larger compound collections using Frog2. The server and documentation are freely available at http://bioserv.rpbs.univ-paris-diderot.fr/Frog2.
INTRODUCTION
Disposing the 3D structure of small drug-like molecules can be critical for several computational approaches such as in silico screening (1,2), either ligand-based (3–5) or receptor structure-based (6–9) employed prior to or to complement experimental screening for hit identification, lead optimization or chemical biology purposes. In addition, for some of the methods, like rigid ligand docking or 3D ligand-based screening, a multiple conformer ensemble is required. Chemical compounds are often distributed by chemical vendors in 1D SMILES (simplified molecular input line entry system), 1D cansmiles (canonical SMILES) (10) or in 2D SDF (11) (structure data file) formats. Generating an accurate 3D structure for a small chemical compound is a complex task (12). Different techniques using rule-based or data-based methods, building linker regions on pre-generated fragments or stochastic procedures up to quantum mechanical methods (12,13) have been developed. Numerous studies have been carried out to compare the existing approaches and to analyze the small molecule conformations experimentally observed (14,15). They revealed that for a satisfactory sampling of the conformational space, the most important parameters to be optimized are the energy window with respect to the global minimum and the root mean square deviation (RSMD) value. Several well established commercial packages such as Corina (Corina Molecular Networks, GmbH Computerchemie Langemarckplatz 1, Erlangen, Germany, 2000), Omega, Catalyst (14) or MED-3DMC (16) generate multiple ensemble conformations of small molecules. In addition, several utilities like Zinc (17), FAF-drugs (18) or pubChem (18) take advantage of commercial software to propose pregenerated collections of compounds in 3D. Yet, very few free tools are available for a single or multiple conformation generation. For instance, Balloon (13) using a multi-objective genetic algorithm approach and Multiconf-DOCK http://dock.compbio.ucsf.edu/Contributed_Code/index.htm (20) using a systematic search are freely available. The open source program DG-AMMOS (21) based on a distance geometry approach and molecular mechanics optimization has been recently reported. A practical alternative of the standalone packages, in particular for non-advanced users, are the web services which can provide direct 1D/2D to 3D facilities, such as OpenEye's Omega, Molsoft, Corina and from some academic sites such as at CBS. Such services, however, usually treat one molecule at a time. Three years ago we developed and reported the web-service Frog (22) (http://bioserv.rpbs.univ-paris-diderot.fr/Frog.html) providing an on-line generation of a single or ensembles of 3D conformations for drug-like compounds. Frog is a mixed rule-based data-based approach based on Frowns (a chemoinformatics toolkit available at http://frowns.sourceforge.net/) to which several functionalities have been added to allow the generation of 3D structures starting from SMILES or SDF data input.
Here, we describe a new version of Frog, Frog2, which is able to (i) generate single or ensembles of low to medium energy 3D conformations starting from 1D/2D or 3D input structure, to (ii) fully or partially disambiguate compound stereochemistry including chiral sites with a user-defined maximum number of generated conformers, and to (iii) minimize the energy of the generated conformers using AMMOS (18) if the user requires. The following important improvements are achieved in Frog2 compared to Frog1: (i) generation of compound rings not available in the fragment database of Frog1 using DG-AMMOS (17); (ii) significantly improved diversity of the generated conformer ensembles, and (iii) considerable increase of the computation speed. In addition, several ancillary tools are provided, such as the format interconversion using OpenBabel (23) or energy minimization of 3D conformations via AMMOS (24). Frog2 still does not allow ring flexibility during the multiple confirmation generation which is under development.
In this study, in addition to describing the web-service Frog2, we validate Frog2 on compounds from the Astex dataset (25). The comparison of Frog2 with Frog1 and the commercial package Omega (http://www.eyesopen.com) shows persuasive performance of Frog2.
METHODS
Concepts
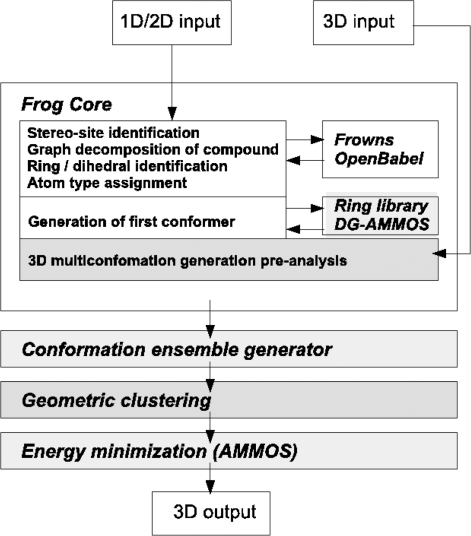
A general overview of the Frog2 service is presented in Figure 1. The new and the optimized modules of Frog2 are shown in the gray white boxes. Overall, the Frog internal 3D generation is based on a graph decomposition of the compound (22) coupled with an identification of the stereo centers for which the chirality is unspecified. Once these sites have been identified, the combinatorial of the unambiguous isomers—for which the chirality of all the identified stereo centers is completely specified—is generated, but randomly truncated to a maximum of eight chiral centers for 3D generation. For each of these, a starting 3D conformation is generated. During this generation, Frog2 takes advantage of DG-AMMOS to generate on the fly missing rings and adds them to the ring library, thus escaping a major limitation of Frog1. Frog does not manage the protonation explicitly. Instead, it relies on OpenBabel (23) to generate hydrogen coordinates for a standard protonation state.
Figure 1.
Flowchart of Frog2.
Another Frog2 major improvement comes from the optimization of the conformation ensemble generation engine. As in Frog1 however, rings are maintained rigid and only dihedral variations are considered. Frog2 engine embeds an improved two stage Monte Carlo procedure. The first stage explores the conformational flexibility based on a limited number of representative dihedral angular values depending on atom types. This Monte-Carlo step includes a procedure to avoid conformation redundancy: new conformations that correspond to previously visited combinations of dihedral values are forbidden. Also, the possibility of biasing the exploration of dihedral values depending on their position relative to the center of the compound has been introduced. For this, the probability that a step affects a particular dihedral is not uniform, but depends on the number of flanking atoms at each side, using the following weight:  , where
, where  (resp.
(resp.  ) is the number of atoms at the left (resp. right) of the bond undergoing the rotation,
) is the number of atoms at the left (resp. right) of the bond undergoing the rotation,  is the total number of atoms, so that dihedrals located at terminal positions have smaller weights compared to dihedrals having a balanced number of atoms each side. The second-stage Monte Carlo uniformly considers small rotations within the stage one conformations, so as to refine them. In order to prune the combinatorial exploration, this stage two is active by default only for conformations of high enough energies.
is the total number of atoms, so that dihedrals located at terminal positions have smaller weights compared to dihedrals having a balanced number of atoms each side. The second-stage Monte Carlo uniformly considers small rotations within the stage one conformations, so as to refine them. In order to prune the combinatorial exploration, this stage two is active by default only for conformations of high enough energies.
This described conformation ensemble generation engine is supplemented by a standard divisive hierarchical clustering using an approach similar in essence to that used for fingerprints (26). This approach guarantees that all conformers in a class are below a fixed RMSD threshold and that the RMSD between selected conformers are always above that threshold.
Finally, in order to speed the computations, the rank of the conformers is based on an internal score of Frog2 taking into account only van der Waals interactions, which might be insufficient to prevent some geometry distortions during the assembly process. To overcome this limitation, it is possible to minimize the conformers using the AMMP force field as implemented in AMMOS (24).
Input/output
Frog2 accepts as input three formats widely used by the community: SMILES, SDF and mol2 formats. It is possible to convert to one of these ones using the OpenBabel facility. Since the SDF and mol2 formats can correspond to both 2D or 3D descriptions of a compound, the user must be precise among the two possible types of input: 1D/2D or 3D. Specifying the 1D/2D type, all 3D information of the input will be discarded and the 3D generation will be performed from scratch. Specifying the 3D type, Frog2 will simply call the conformation ensemble generation engine, not considering stereoisomery and taking the input coordinates as a starting point for the conformation ensemble generation. Parameters related to the generation of ensembles correspond to a maximal number of conformations, and energy thresholds to define the allowed energy window referred to the lowest energy conformation generated. It is also possible to specify a minimal RMSD value for geometric clustering of the conformers, and to invoke the minimization of the compounds. The minimization is only applied on exit of the generation. The energy minimization takes into account the valence, angle and van der Waals components, not considering electrostatics terms. The internal format for the Frog2 results is mol2. It is, however, possible to choose between mol2, SDF or PDB formats, thanks to OpenBabel.
RESULTS
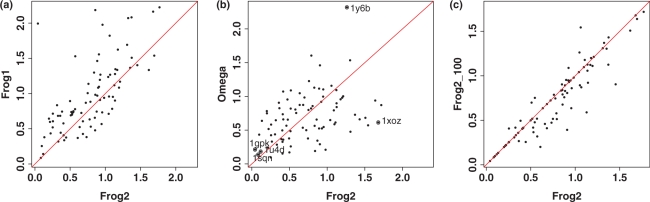
We first assessed the Frog2 performance for finding conformation similar to bioactive ones among the generated multiconformation ensemble. Figure 2 shows the results for generating multiconformation ensembles of the Astex dataset (25) containing 85 diverse drug-like molecules using Frog2, Frog1 and Omega 2 [Openeye Scientific Software (http://www.eyesopen.com)]. The same input parameters were applied to run Frog2, Frog1 and Omega, namely: up to 50 conformers, RMSD threshold of 0.8.Å. In order to make possible the comparison with Frog1, the Frog2 disambiguation option was activated. As can be seen from Figure 2a, using these input parameters, applying unambiguation for the stereoisomery and allowing the stage two Monte-Carlo, Frog2 finds conformations closer to the bioactive ones than Frog1 for most of the Astex molecules. On average, the Frog2 bioactive' closest conformation is at  Å RMSD values whereas Frog1 shows an average RMSD of
Å RMSD values whereas Frog1 shows an average RMSD of  Å. Interestingly, Frog2 performs better than Frog1 even without employing the stage two Monte Carlo with an average RMSD of
Å. Interestingly, Frog2 performs better than Frog1 even without employing the stage two Monte Carlo with an average RMSD of  , respectively. Similar results in terms of RMSD are obtained using Frog2 and Omega when a maximum of 50 conformers were generated on the Astex dataset with a slight, outperformance of Omega with average values being of
, respectively. Similar results in terms of RMSD are obtained using Frog2 and Omega when a maximum of 50 conformers were generated on the Astex dataset with a slight, outperformance of Omega with average values being of  (Wilcoxon signed rank test shows no significant difference). For a maximum of 50 conformers per molecule, Omega and Frog2 found conformations within an RMSD with the bioactive one of 1.5 Å for 82 and 79 compounds out of 85, respectively. That can be considered as an acceptable accuracy keeping in mind that the required RMSD for clustering the similar conformations was set to 0.8 Å. Finally, Figure 2c illustrates the impact of increasing the maximal number of conformers up to 100. One notes a small, but effective improvement. On average, Frog2 finds the bioactive conformation within
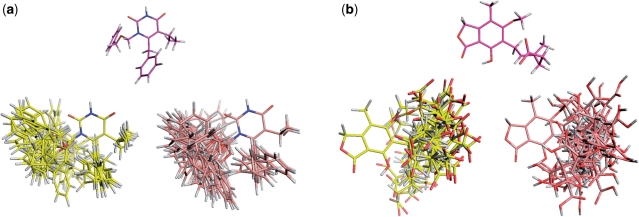
(Wilcoxon signed rank test shows no significant difference). For a maximum of 50 conformers per molecule, Omega and Frog2 found conformations within an RMSD with the bioactive one of 1.5 Å for 82 and 79 compounds out of 85, respectively. That can be considered as an acceptable accuracy keeping in mind that the required RMSD for clustering the similar conformations was set to 0.8 Å. Finally, Figure 2c illustrates the impact of increasing the maximal number of conformers up to 100. One notes a small, but effective improvement. On average, Frog2 finds the bioactive conformation within  Å RMSD. Examples demonstrating the conformational diversity achieved by Frog2 and Omega when 50 conformers generated for two molecules of the Astex dataset can be seen in Figure 3. According to the computed RMSD values and visual analysis, one can conclude that both Frog2 and Omega explore quite well the conformational space and are able to generate conformations that are close to the bioactive one. In addition, the user has the possibility to energy-minimize the generated conformers via Frog2 or to minimize a own compound library in 3D. Our assessment of the impact of the minimization on the Frog2-generated Astex conformers does not show a significant improvement by means of finding the bioactive conformations. On average, the RMSD from the bioactive conformations was of
Å RMSD. Examples demonstrating the conformational diversity achieved by Frog2 and Omega when 50 conformers generated for two molecules of the Astex dataset can be seen in Figure 3. According to the computed RMSD values and visual analysis, one can conclude that both Frog2 and Omega explore quite well the conformational space and are able to generate conformations that are close to the bioactive one. In addition, the user has the possibility to energy-minimize the generated conformers via Frog2 or to minimize a own compound library in 3D. Our assessment of the impact of the minimization on the Frog2-generated Astex conformers does not show a significant improvement by means of finding the bioactive conformations. On average, the RMSD from the bioactive conformations was of  Å, i.e. similar results to the ones obtained by Frog2 without a final minimization stage. In addition, we validated the Frog2 performance on a larger collection of compounds taken from the PDBbind database (27). We extracted all PDBbind ligands with an X-ray resolution better than 2Å. The compounds contatining up to 15 rotatable bonds and not including large bridged rings (more than 30 atoms, see the discussion about Frog2 limitations above) were retained. Finally, 962 compounds were treated. The distribution of the resulted test-set compounds depending on the number of rotatable bonds is given in the Supplementary Data. The results for RMSD between the best generated and experimental structures are also given in the Supplementary Data. The conformer ensembles were generated for a maximum of 50 conformers, enabling stage two Monte Carlo, and without applying minimization. As can be expected, the performance (in terms of RMSD) decreases with increasing molecular flexibility. The median RMSD to the experimental conformation is below 1Å up to 7 rotatable bonds, and Frog2 results remain however acceptable up to 15 rotatable bonds.
Å, i.e. similar results to the ones obtained by Frog2 without a final minimization stage. In addition, we validated the Frog2 performance on a larger collection of compounds taken from the PDBbind database (27). We extracted all PDBbind ligands with an X-ray resolution better than 2Å. The compounds contatining up to 15 rotatable bonds and not including large bridged rings (more than 30 atoms, see the discussion about Frog2 limitations above) were retained. Finally, 962 compounds were treated. The distribution of the resulted test-set compounds depending on the number of rotatable bonds is given in the Supplementary Data. The results for RMSD between the best generated and experimental structures are also given in the Supplementary Data. The conformer ensembles were generated for a maximum of 50 conformers, enabling stage two Monte Carlo, and without applying minimization. As can be expected, the performance (in terms of RMSD) decreases with increasing molecular flexibility. The median RMSD to the experimental conformation is below 1Å up to 7 rotatable bonds, and Frog2 results remain however acceptable up to 15 rotatable bonds.
Figure 2.
RMSD between the best-fitted conformers and the X-ray structures for conformers generated by: (a) Frog1 versus Frog2 for series of up to 50 conformers; (b) Frog2 versus Omega for series of up to 50 conformers. PDB codes denote (i) the compounds with poorest relative performance, i.e. deviating the most from the diagonal (1y6b, 1xoz) and (ii) best performing for both Frog2 and Omega (1gpk, 1u4d, 1sqn); and (c) Frog2 for series of up to 50 conformers versus Frog2 for series of up to 100 conformers.
Figure 3.
Conformational ensembles for two small molecules of the Astex dataset generated by Frog2 (all atom colors, carbons in yellow) and Omega (all atom colors, carbons in light pink: (a) PDB code 1jla; and (b) PDB code 1meh. The experimental structure of the co-crystallized ligands are also shown (all atom colors, carbons in magenta).
Finally, we assessed the gain in speed of Frog2 compared to Frog1. On a computer with intel Xeon processors at 2 GHZ, Omega-generated conformers for all the Astex set in 6 min. The generation using Frog2 required 11 min. Using comparable parameters, Frog1 generated the same collection in 103 min. In addition, the computational time can be reduced to 5 min if Frog2 is run without the two Monte Carlo stage. In summary, Frog2 generates better conformational quality and diversity than Frog1 and is nine times faster than Frog1 using conditions strictly comparable. Frog2 still outperforms Frog1 when employing the faster approach without two-stage Monte Carlo in terms of conformational diversity and is 20 times faster. This dramatic decrease in execution time without affecting the quality of the generated conformations demonstrates the impact of the optimizations implemented into Frog2. We should note that activating the minimization option leads to significant increase of the computational time. Over the Astex set, using the disambiguation option and stage two Monte Carlo, the calculation times increased from 11 min up to 207 min, i.e. close to 20 times slower. However, the minimization may be required to energy-minimize some generated structures with detected structural inaccuracies, or for a small number of compounds.
DISCUSSION AND FUTURE DIRECTIONS
The main goal of the Frog2 development was to overcome several limitations of Frog1 and to increase the overall quality and speed performance. Over these, rings were a central concern. Indeed, Frog2 dependency on a ring library is much less critical since Frog2 takes the advantage of DG-AMMOS to generate on the fly missing rings. Furthermore, Frog2 addresses a Frog1 issue problem related to rings' protonation now generated standardly via OpenBabel. As a result, the Frog2 failure rate to generate conformers is low. Used over 10 000 compounds randomly selected out of ∼40 0000 cmps from the ChemBrdge diverset (http://www.chembridge.com/), Frog2 only failed to generate conformers for <2% of the compounds. As illustrated from the tests on the Astex set, Frog2 reaches a good structure quality.
A second focus concerned the conformational diversity. The Frog2 development permitted to better control the diversity in the conformation ensemble generation and Frog2 reaches extensive conformational diversity. Further improvements are expected. In particular, better consideration of symmetry is under investigation.
Last focus was related to needed computational time. Indeed, in a context of virtual screening experiments, where millions of compounds can be considered, the time to generate libraries can become a concern. One can suppose that once generated in 3D, a chemical compound collection can be in silico screened for different projects, however, more and more generation of focused libraries, either target-based or ligand-based, is required to increase the efficiency of discovery programs. In this respect, Frog2 brings significant improvement since it is able to generate satisfactory quality conformations 20 times faster than Frog1. We should note that a balance between the speed and conformational diversity can be achieved depending on the project purposes and the number of compounds to treat. Inactivating the stage two Monte Carlo, operational on the server, will result in a faster generation, but a reduced conformational diversity will be reached. For applications requiring best explored conformational space, it is thus recommended to maintain this stage active. Finally, Frog2 takes advantage of AMMOS to offer the possibility to minimize the conformations that can be very helpful in some particular cases, even its use results in dramatically larger computational cost.
Several limitations are still present in Frog2. The first one is related to the ring rigidity that can affect the quality or the diversity of the conformations generated in some cases. Particularly, compounds including large bridged rings for which flexibility impacts the conformational search are presently out of Frog scope. Also, a side effect of the ring library strategy is that Frog2 is presently not able to treat properly some cases of stereoisomery involving stereo centers in rings. Finally, some errors in the generation of some particular groups can be presently observed since Frog2 builds from scratch the linkers, at a single atom level, i.e. not combining pregenerated fragments. Efforts will now focus on the implementation of specific rules for such groups. In addition, the implementation of some ring flexibility is under development.
Despite of the minor current limitations, the assessment of Frog2 demonstrated a convincing performance. The use of Frog2 could help for various in silico studies, from ligand-based or structure-based virtual screening, to lead compounds optimization etc. Thus, the Frog2 server offers a very valuable tool which provides efficient features to the community for an extremely competitive computational time.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Funding for open access charge: INSERM UMR-S 973 recurrent funding.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors thank O. Sperandio, M. Petitjean, D. Lagorce and J. Maupetit for many useful discussion and suggestions.
REFERENCES
- 1.Shoichet BK. Virtual screening of chemical libraries. Nature. 2004;432:862–865. doi: 10.1038/nature03197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Clark D. What has virtual screening ever done for drug discovery? Expert. Opin. Drug. Discov. 2008;3:841–851. doi: 10.1517/17460441.3.8.841. [DOI] [PubMed] [Google Scholar]
- 3.Douguet D. Ligand-based approaches in virtual screening. Cur. Comput. Aided Drug Des. 2008;4:180–190. [Google Scholar]
- 4.Pajeva IK, Globisch C, Wiese M. Combined pharmacophore modeling, docking, and 3D QSAR studies of ABCB1 and ABCC1 transporter inhibitors. ChemMedChem. 2009;4:1883–1896. doi: 10.1002/cmdc.200900282. [DOI] [PubMed] [Google Scholar]
- 5.Bender A, Jenkins JL, Scheiber J, Sukuru S.CK, Glick M, Davies JW. How similar are similarity searching methods? A principal component analysis of molecular descriptor space. J. Chem. Inf. Model. 2009;49:108–119. doi: 10.1021/ci800249s. [DOI] [PubMed] [Google Scholar]
- 6.Kuntz ID. Structure-based strategies for drug design and discovery. Science. 1992;257:1078–1082. doi: 10.1126/science.257.5073.1078. [DOI] [PubMed] [Google Scholar]
- 7.Villoutreix BO, Renault R, Lagorce D, Sperandio O, Montes M, Miteva MA. Free resources to assist structure-based virtual ligand screening experiments. Curr. Protein Pept. Sci. 2007;8:381–411. doi: 10.2174/138920307781369391. [DOI] [PubMed] [Google Scholar]
- 8.Betzi S, Restouin A, Opi S, Arold ST, Parrot I, Guerlesquin F, Morelli X, Collette Y. Protein protein interaction inhibition (2P2I) combining high throughput and virtual screening: application to the HIV-1 Nef protein. Proc. Natl Acad. Sci. USA. 2007;104:19256–19261. doi: 10.1073/pnas.0707130104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Seifert MHJ. Targeted scoring functions for virtual screening. Drug Discov. Today. 2009;14:562–569. doi: 10.1016/j.drudis.2009.03.013. [DOI] [PubMed] [Google Scholar]
- 10.Weininger D, Weininger A, Weininger JL. SMILES. 2. Algorithm for generation of unique SMILES notation. J. Chem. Inf. Comput. Sci. 1989;29:97–101. [Google Scholar]
- 11.Dalby A, Nourse JG, Hounshell WD, Gushurst AK, Grier DL, Leland BA, Laufer J. Description of several chemical structure file formats used by computer programs developed at molecular design limited. J. Chem. Inf. Comput. Sci. 1992;32:244–255. [Google Scholar]
- 12.Sadowski J, Gasteiger J. From atoms and bonds to three-dimensional atomic coordinates: automatic model builders. Chem. Rev. 1993;93:2567–2581. [Google Scholar]
- 13.Vainio MJ, Johnson MS. Generating conformer ensembles using a multiobjective genetic algorithm. J. Chem. Inf. Model. 2007;47:2462–2474. doi: 10.1021/ci6005646. [DOI] [PubMed] [Google Scholar]
- 14.Kirchmair J, Wolber G, Laggner C, Langer T. Comparative performance assessment of the conformational model generators omega and catalyst: a large-scale survey on the retrieval of protein-bound ligand conformations. J. Chem. Inf. Model. 2006;46:1848–1861. doi: 10.1021/ci060084g. [DOI] [PubMed] [Google Scholar]
- 15.Brameld KA, Kuhn B, Reuter DC, Stahl M. Small molecule conformational preferences derived from crystal structure data. A medicinal chemistry focused analysis. J. Chem. Inf. Model. 2008;48:1–24. doi: 10.1021/ci7002494. [DOI] [PubMed] [Google Scholar]
- 16.Sperandio O, Souaille M, Delfaud F, Miteva MA, Villoutreix BO. MED-3DMC: a new tool to generate 3D conformation ensembles of small molecules with a Monte Carlo sampling of the conformational space. Eur. J. Med. Chem. 2009;44:1405–1409. doi: 10.1016/j.ejmech.2008.09.052. [DOI] [PubMed] [Google Scholar]
- 17.Irwin JJ, Shoichet BK. ZINC–a free database of commercially available compounds for virtual screening. J. Chem. Inf. Model. 2005;45:177–182. doi: 10.1021/ci049714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Miteva MA, Violas S, Montes M, Gomez D, Tuffery P, Villoutreix BO. FAF-drugs: free adme/tox filtering of compound collections. Nucleic Acids Res. 2006;34:W738–W744. doi: 10.1093/nar/gkl065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wheeler DL, Barrett T, Benson DA, Bryant SH, Canese K, Chetvernin V, Church DK, DiCuccio M, Edgar R, Federhen S, et al. Database resources of the national center for biotechnology information. Nucleic Acids Res. 2007;35:D5–D12. doi: 10.1093/nar/gkl1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sauton N, Lagorce D, Villoutreix BO, Miteva MA. MS-DOCK: accurate multiple conformation generator and rigid docking protocol for multi-step virtual ligand screening. BMC Bioinformatics. 2008;9:184. doi: 10.1186/1471-2105-9-184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lagorce D, Pencheva R, Villoutreix BO, Miteva MA. Dg-ammos: a new tool to generate 3D conformation of small molecules using distance geometry and automated molecular mechanics optimization for in silico screening. BMC Chem Biol. 2009;9:6. doi: 10.1186/1472-6769-9-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bohme Leite T, Gomes D, Miteva MA, Chomilier J, Villoutreix BO, Tufféry P. Frog: a free online drug 3D conformation generator. Nucleic Acids Res. 2007;35:W568–W572. doi: 10.1093/nar/gkm289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guha R, Howard MT, Hutchison GR, Murray-Rust P, Rzepa H, Steinbeck C, Wegner J, Willighagen EL. The blue obelisk-interoperability in chemical informatics. J. Chem. Inf. Model. 2006;46:991–998. doi: 10.1021/ci050400b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pencheva T, Lagorce D, Pajeva I, Villoutreix BO, Miteva MA. Ammos: automated molecular mechanics optimization tool for in silico screening. BMC Bioinformatics. 2008;9:438. doi: 10.1186/1471-2105-9-438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hartshorn MJ, Verdonk ML, Chessari G, Brewerton SC, Mooij WTM, Mortenson PN, Murray CW. Diverse, high-quality test set for the validation of protein-ligand docking performance. J. Med. Chem. 2007;50:726–741. doi: 10.1021/jm061277y. [DOI] [PubMed] [Google Scholar]
- 26.Haigh JA, Pickup BT, Grant JA, Nicholls A. Small molecule shape-fingerprints. J. Chem. Inf. Model. 2005;45:673–684. doi: 10.1021/ci049651v. [DOI] [PubMed] [Google Scholar]
- 27.Wang R, Fang X, Lu Y, Wang S. The PDBbind database: collection of binding affinities for protein-ligand complexes with known three-dimensional structures. J. Med. Chem. 2004;47:2977–2980. doi: 10.1021/jm030580l. [DOI] [PubMed] [Google Scholar]