Abstract
The EMBL-EBI provides access to various mainstream sequence analysis applications. These include sequence similarity search services such as BLAST, FASTA, InterProScan and multiple sequence alignment tools such as ClustalW, T-Coffee and MUSCLE. Through the sequence similarity search services, the users can search mainstream sequence databases such as EMBL-Bank and UniProt, and more than 2000 completed genomes and proteomes. We present here a new framework aimed at both novice as well as expert users that exposes novel methods of obtaining annotations and visualizing sequence analysis results through one uniform and consistent interface. These services are available over the web and via Web Services interfaces for users who require systematic access or want to interface with customized pipe-lines and workflows using common programming languages. The framework features novel result visualizations and integration of domain and functional predictions for protein database searches. It is available at http://www.ebi.ac.uk/Tools/sss for sequence similarity searches and at http://www.ebi.ac.uk/Tools/msa for multiple sequence alignments.
INTRODUCTION
Bioinformatics is a vast and complex multidisciplinary research area where numerous tools have been developed over the years to analyse constantly growing amounts of data. Since 1998, the European Bioinformatics Institute (EMBL–EBI) has provided public access to various mainstream sequence analysis applications (1,2). These include sequence similarity search services (http://www.ebi.ac.uk/Tools/similarity.html), such as FASTA (3), BLAST (4,5) and InterProScan (6) and multiple sequence alignment tools (http://www.ebi.ac.uk/Tools/sequence.html), such as ClustalW (7), T-Coffee (8), MUSCLE (9), Kalign (10) and MAFFT (11). These services are provided via a PERL-CGI job dispatcher framework for managing job submission and result representation. This infrastructure handled more than 16 million jobs during 2009. The popularity of these services has made it necessary to redesign the system in order to minimize maintenance and enhance the integration of features requested by users. A new and modular framework, called JDispatcher, has been developed to improve the accessibility and quality of the services relevant to the biological community.
JDispatcher framework
JDispatcher is aimed at both novice and expert users and exposes novel methods of obtaining annotations and visualizing sequence analysis results through one uniform and consistent interface. These services are available interactively over the web and via SOAP and REST interfaces for systematic or programmatic use. The new framework provides input validation to assure successful job submissions, offers new visualization features to assist in the interpretation of results and uses the EBI search engine, EB-eye (12), to integrate relevant annotations.
A user can submit sequences using web forms that contain all supported parameters and their possible values. The different tools have been grouped into categories based on their purpose (Table 1).
Table 1.
Tools available in the JDispatcher framework
| Category | Tool |
|---|---|
| Sequence Similarity Searches (sss) | psisearch, psiblast, ncbiblast, wublast, fasta, ssearch, ggsearch and glsearch |
| Multiple Sequence Alignments (msa) | clustalw2, tcoffee, kalign, muscle, mafft, and prank |
Within a category, the tools share the same interface design, which uses well established usability patterns, such as wizard-like steps to guide the user through the submission process. It makes use of decision-trees to validate all the parameters required to warrant successful job submissions. If the validation fails, the user is notified about which specific parameters or data are invalid, and the job is not submitted. Alternatively, JDispatcher assigns a unique job identifier and sends a request to a workload management system for the job to be executed. The identifier is then used to keep track of the tasks and to retrieve the results when they become available. The results of each job are kept for a maximum of 7 days.
Results representation
The results of an analysis are made available using various representations (e.g. HTML tables, XML files, images, etc.). In order to produce these representations, each result is converted into a generic category-specific model that is used by a renderer that generates the requested output. The renderers are specific to the model and not to the tool, and thus are available across all the tools in a category. The availability of multiple views of the same data helps the user to interpret and compare results from different tools within a category.
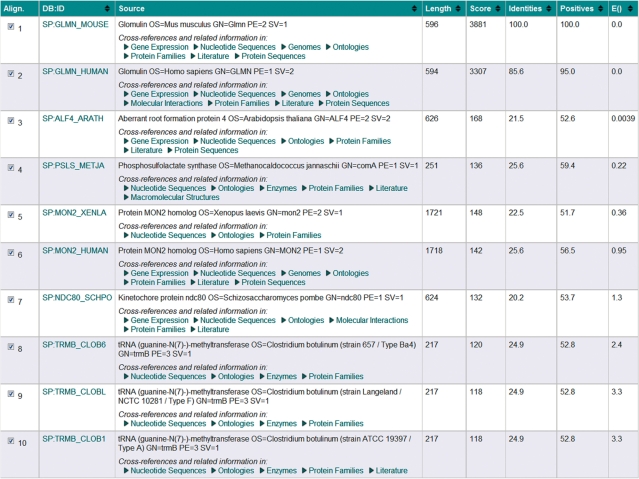
Sequence search algorithms produce limited hits annotation. With the new framework it is possible to navigate hits and access related information. Figure 1 shows the ‘Summary Table’ of an SSEARCH of mouse glomulin (UniProtKB/Swiss-Prot GLMN_MOUSE), which is essential for the development of the vascular system, against the UniProtKB/Swiss-Prot database (13). Each column heading has clickable arrows that allow the user to sort the results according to the values in the columns [e.g. sequence length, score, percentage identity, positives and E()-value]. Each match is enriched with links to cross-references and related information in various data resources (e.g. gene expression, genomic sequences, structures, function, ontologies and literature citations). Optionally, the alignment from the search, and/or the full-annotation for the selected matches can be displayed. A hits selection can also be downloaded in fasta format.
Figure 1.
Summary Table view of the results obtained when searching the sequence of mouse glomulin against the UniProtKB/Swiss-Prot database using SSEARCH.
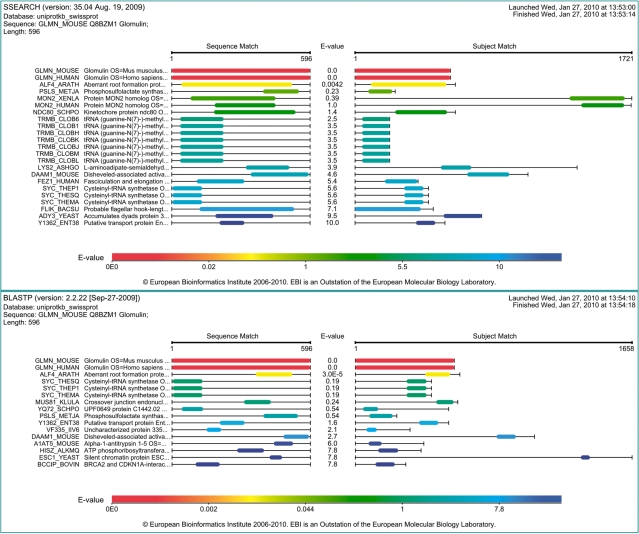
Figure 2 shows the ‘Visual Output’ obtained from searches using SSEARCH and NCBI BLAST of the glomulin sequence against UniProtKB/Swiss-Prot using default parameters. Comparison of the two images reveals notable differences in the sequence matches reported by the two search methods. For example, differences in the aligned regions between glomulin and aberrant root formation protein 4 for Arabidopsis (ALF4_ARATH) are clearly visible in both; SSEARCH identifies two MON2 homologues at E()-values <1 (MON2_XENLA and MON2_HUMAN), which may indicate there is a structural relationship between GLMN at the C-terminus of the MON2 homologues, although these may not share related functions.
Figure 2.
Comparisons between the Visual output results obtained when searching the sequence of mouse glomulin against the UniProtKB/Swiss-Prot database using SSEARCH and NCBI BLAST, respectively.
Determining which functional domains and families a protein belongs to is critical to the understanding of the biological processes it may be involved in. This is important for the characterization of existing drug targets as well as in the identification of novel ones. Family and domain functional predictions have been built into the framework, using pre-calculated matches from the InterPro Consortium (14) data. This enables users, not only to search for sequence similarities when using the UniProt databases, but also to characterize the sequence query in terms of domain architectures that may elicit its function. Figure 3 shows ‘Functional Predictions’ for a hypothetical bioactive lysophospholipid that was compared against UniProtKB/Swiss-Prot using NCBI BLAST. The hypothetical sequence has several good homologues, all belonging to the GPCR rhodopsin-like superfamily, which are clearly seen. This indicates the query protein could represent a potential target for receptor-binding studies.
Figure 3.
Functional prediction view of the results obtained when comparing the sequence of putative bioactive lysophospholipid that was compared against UniProtKB/Swiss-Prot using NCBI BLAST.
In both, the ‘Visual Output’ and ‘Functional Predictions’ result representations, the matches are coloured, from red to blue, according to E()-value, using a relative scale, from the most to the least significant hits within the result. An absolute scale, which ranges from E() = 0 to E()=1.0, is also available. These aim to aid the user in deciding whether weak similarities may be biologically significant. These images are available in Scalable Vector Graphics (SVG), Portable Network Graphic (PNG) and JPEG output, providing wide compatibility. The raw result and processed forms, such as the ‘Summary Table’ content and XML formats are downloadable for further processing by the user.
The examples above illustrate how, from a single sequence similarity search, it is possible to access related sources of annotation, determine visually which results are relevant and infer gene and protein functional associations, using the JDispatcher framework.
Web Services
Web Services technologies have opened up important opportunities for the analysis of life sciences data. It is now well established that sharing resources, across geographically distributed networks, is advantageous to scientists and bioinformaticians through the re-use of generic services, such as those presented in this article. The new JDispatcher framework provides multiple front-ends: in addition to the web interface, SOAP and REST APIs (http://www.ebi.ac.uk/Tools/webservices/) have been implemented to offer programmatic access using accepted web services standards.
The SOAP and REST APIs cater for users requiring systematic access to a wide range of sequence similarity search and multiple sequence alignment services, which can be built into local analytical workflows and pipelines (e.g. Taverna (15), Triana (http://www.trianacode.org/), KNIME (www.knime.org) (16) and Pipeline Pilot (http://accelrys.com/products/scitegic/index.html))—typical usage scenarios include the characterization of novel genomes and proteomes and the analysis of data derived from meta-genome experiments.
Using the APIs, complex applications can be developed in various programming languages, which include: C/C++, C#, Java, Perl, PHP, Python and Ruby, or scripting environments such a Bash, csh, batch and PowerShell. This allows integration of services into existing and/or new applications that require access to fast sequence database searching or multiple sequence alignment methods. To facilitate this type of usage, the services provide extensive meta-information describing the available parameters, including their possible values and descriptions of their purpose.
Typical applications of the JDispatcher framework services include: providing an alternative interface for specialist usage targeted at a specific community; integrating a service into an existing data portal to provide analysis services; and enhancing analysis results by directly connecting the result with the data. These are of importance to service providers and users of pipelines who may not have the resources to run and maintain the infrastructure required to support equivalent functionality.
CONCLUSIONS
The modularity of this new framework reduces maintenance overheads and simplifies the addition of tools and features. Keeping the result data model and the renderers separate provides the flexibility to add additional representations to all functionally related tools. This improves the level of usability for both novice and expert users. The presented visualization examples highlight important insights in the understanding of existing and new nucleotide and protein sequences from both genomes and metagenome experiments and suggest novel ways in which these data can be interpreted.
Academic and commercial laboratories can integrate the JDispatcher framework services with their local analytical pipelines or workflows. These represent an important contribution to the growing number of available services in bioinformatics and have been submitted to the BioCatalogue (17) (www.biocatalogue.org), a registry of freely available web services in the life sciences.
FUNDING
The European Commission under FELICS [contract number 021902 (RII3), within the Research Infrastructure Action of the FP6 ‘Structuring the European Research Area’ Programme]; core funding from the European Molecular Biology Laboratory; European Patent Office. Funding for open access charge: EMBL.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We acknowledge valuable feedback from Prof. William Pearson from the University of Virginia, USA and the InterPro and UniProt teams at EMBL-EBI.
REFERENCES
- 1.McWilliam H, Valentin F, Goujon M, Li W, Narayanasamy M, Martin J, Miyar T, Lopez R. Web services at the European Bioinformatics Institute—2009. Nucleic Acids Res. 2009;37:W6–W10. doi: 10.1093/nar/gkp302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brooksbank C, Cameron G, Thornton J. The European Bioinformatics Institute’s data resources. Nucleic Acids Res. 2010;38:D17–D25. doi: 10.1093/nar/gkp986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pearson WR, Lipman DJ. Improved tools for biological sequence comparison. Proc. Natl Acad. Sci. USA. 1988;85:2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lopez R, Silventoinen V, Robinson S, Kibria A, Gish W. WU-Blast2 server at the European Bioinformatics Institute. Nucleic Acids Res. 2003;31:3795–3798. doi: 10.1093/nar/gkg573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Quevillon E, Silventoinen V, Pillai S, Harte N, Mulder N, Apweiler R, Lopez R. InterProScan: protein domains identifier. Nucleic Acids Res. 2005;33:W116–W120. doi: 10.1093/nar/gki442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, et al. ClustalW2 and ClustalX version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 8.Notredame C, Higgins D, Heringa J. T-Coffee: a novel method for multiple sequence alignments. J. Mol. Biol. 2000;302:205–217. doi: 10.1006/jmbi.2000.4042. [DOI] [PubMed] [Google Scholar]
- 9.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lassmann T, Sonnhammer EL. Kalign – an accurate and fast multiple sequence alignment algorithm. BMC Bioinformatics. 2005;6:298. doi: 10.1186/1471-2105-6-298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Katoh K, Asimenos G, Toh H. Multiple alignment of DNA sequences with MAFFT. Methods Mol. Biol. 2009;537:39–64. doi: 10.1007/978-1-59745-251-9_3. [DOI] [PubMed] [Google Scholar]
- 12.Valentin F, Squizzato S, Goujon M, McWilliam H, Paern J, Lopez R. Fast and efficient searching of biological data resources—using EB-eye. Brief. Bioinformatics. 2010 doi: 10.1093/bib/bbp065. doi:10.1098/bib/bbp065 [Epub ahead of print 11 February 2010] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.The UniProt Consortium. The Universal Protein Resource (UniProt) in 2010. Nucleic Acids Res. 2010;38:D142–D148. doi: 10.1093/nar/gkp846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hunter S, Apweiler R, Attwood TK, Bairoch A, Bateman A, Binns D, Bork P, Das U, Daugherty L, Duquenne L, et al. InterPro: the integrative protein signature database. Nucleic Acids Res. 2009;37:D211–D215. doi: 10.1093/nar/gkn785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hull D, Wolstencroft K, Stevens R, Goble C, Pocock MR, Li P, Oinn T. Taverna: a tool for building and running workflows of services. Nucleic Acids Res. 2006;34:W729–W732. doi: 10.1093/nar/gkl320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Berthold MR, Cebron N, Dill F, Gabriel TR, Kotter T, Mein T, Ohl P, Sieb C, Thiel K, Wiswedel B. Data Analysis, Machine Learning and Applications – Proceedings of the 31st Annual Conference of the Gesellschaft für Klassifikation e.V., Studies in Classification, Data Analysis, and Knowledge Organization. Berlin, Germany: Springer; 2007. KNIME: The Konstanz Information Miner; pp. 319–326. [Google Scholar]
- 17.Goble C, Belhajjame K, Tanoh F, Bhagat J, Wolstencroft K, Stevens R, Nzuobontane E, McWilliam H, Laurent T, Lopez R. BioCatalogue: a curated web service registry for the life science community. Nature Precedings. 2009 http://www.iscb.org/uploaded/css/36/11627.pdf. [Google Scholar]