Abstract
mGenomeSubtractor performs an mpiBLAST-based comparison of reference bacterial genomes against multiple user-selected genomes for investigation of strain variable accessory regions. With parallel computing architecture, mGenomeSubtractor is able to run rapid BLAST searches of the segmented reference genome against multiple subject genomes at the DNA or amino acid level within a minute. In addition to comparison of protein coding sequences, the highly flexible sliding window-based genome fragmentation approach offered can be used to identify short unique sequences within or between genes. mGenomeSubtractor provides powerful schematic outputs for exploration of identified core and accessory regions, including searches against databases of mobile genetic elements, virulence factors or bacterial essential genes, examination of G+C content and binucleotide distribution bias, and integrated primer design tools. mGenomeSubtractor also allows for the ready definition of species-specific gene pools based on available genomes. Pan-genomic arrays can be easily developed using the efficient oligonucleotide design tool. This simple high-throughput in silico ‘subtractive hybridization’ analytical tool will support the rapidly escalating number of comparative bacterial genomics studies aimed at defining genomic biomarkers of evolutionary lineage, phenotype, pathotype, environmental adaptation and/or disease-association of diverse bacterial species. mGenomeSubtractor is freely available to all users without any login requirement at: http://bioinfo-mml.sjtu.edu.cn/mGS/.
INTRODUCTION
Bioinformatics- and microarray-facilitated comparative analyses have revealed that some bacterial species possess extremely plastic genomes, hence giving rise at a population level to the concept of a species-associated meta- or pan-genome (1,2). The pan-genome comprises the entire set of DNA sequences that are represented invariably, frequently, sporadically or only rarely in members of the species. At an individual level the genome of each bacterium can be divided into core regions, shared by the entire species, and accessory or species-variable regions. This latter optional genomic repertoire, termed by us as the ‘mobilome’ (mobile genome) (3), includes a myriad of short strain-specific sequences and longer spans of episomal and integrative plasmids, transposons, integrons, gene cassettes, prophages, strain-specific gene clusters of unknown provenance and a growing list of genomic islands (4,5).
Many recently developed in silico comparison tools, web-servers and databases aim to exploit the expanding flow of high-throughput genome sequencing data to reveal the wider DNA blueprints of individual bacterial species and map out the extent and nature of species-associated mobilomes. The following examples are of particular note: BLAST-based webACT can be used to visualize pair-wide similarity across up to five user-supplied genome sequences (6), the CGview server can generate up to three circular genome maps with user-uploaded BLAST hits (7), MUMmer rapidly aligns a complete or partially sequenced genome against a reference template (8), Mauve functions as a genome-scale multiple sequence aligner (9) and the xBASE database offers a multi-functional comparative analysis resource for laboratory-based bacteriologists (10). GenomeSubtractor 1.0 is an in silico substractive hybridization tool, available as accessory within the tRNAcc command line package (11) and the MobilomeFINDER server (12). We first used this tool to identify protein coding sequences that were present in Salmonella enterica serovar Paratyphi A but not available non-Paratyphi A Salmonella genomes to develop a Paratyphi A-specific multiplex PCR assay (13). However, current freely available comparative genomics tools exhibit two major deficiencies: (i) an inability to analyse several genome sequences at once, especially when the genomes compared are relatively divergent and (ii) excessively long computing times and limited computational outputs as processing is typically performed on a local PC. We have now developed mGenomeSubtractor, a markedly superior version of GenomeSubtractor 1.0, to perform highly parallel genome-scale in silico ‘subtractive hybridization’ analyses of reference genomes against up to forty comparator genomes simultaneously. This mpiBLAST-based tool allows rapid alignment of tens of NCBI-archived or user-supplied genome sequences within a minute. mGenomeSubtractor is a highly intuitive cluster node architecture-based computing tool that offers efficient visualization of genomic mosaics, zoom-in options, a wide range of targeted analyses, linkages to other databases and a useful range of applications. mGenomeSubtractor will be ideal for the identification and exploration of the most dynamic regions of bacterial genomes and for the definition and preliminary characterization of wider species-associated gene pools.
ANALYSIS TOOLS
mGenomeSubtractor web-interface: a mpiBLAST-based tool for parallel in silico subtractive hybridization of bacterial genomes
Using an mpiBLAST-based procedure, mGenomeSubtractor performs a simple and intuitive in silico ‘subtractive hybridization’ by comparing selected closely related genomes to generate a list of conserved/strain-specific protein coding sequences (CDS) of the query genome and subsequently obtain the core/accessory regions. To examine the degree of sequence similarity at a nucleotide or amino acid level between each query CDS and the set of subject genomes, mGenomeSubtractor employs standard BLAST-derived identity and ratio of matching length to query length cut-offs or the simple H-value homology score (3,14). For each query, the H-value was calculated as follows: H = i × (lm/lq), where i was the level of identity of the region with the highest Bit score expressed as a frequency of between 0 and 1, lm the length of the highest scoring matching sequence (including gaps) and lq the query length. In this study, the H-value was used to measure BLASTN-derived sequence similarity at nucleotide level. By analogy, we defined an equivalent Ha-value that reflected BLASTP-based similarity at an amino acid level.
mGenomeSubtractor provides a flexible and biologist-friendly web-interface, consisting of an input page, a retrieve and Results pages. Users select a reference (or query) genome and a set of subject genomes for comparison from a full listing of complete bacterial genomes, regularly updated by NCBI Microbial Genome Resources. In the example shown in Figure 1, the Streptomyces coelicolor A3(2) genome was selected as the reference genome, with the subject set comprising two genomes: the S. avermitilis MA-4680 genome selected from the NCBI genome listing and the complete S. lividans TK24 genome downloaded from the Broad Institute and user-uploaded to the mGenomeSubtractor website. Parallel implementation of mpiBLASTP was used to measure amino acid sequence similarity with default NCBI BLASTP parameters, with the exception that ‘F’ was set to ‘F (no filter for repeated sequences)’. mGenomeSubtractor allows users to select or upload complete sequence and annotation details of reference (and comparator) genomes. Users can also simultaneously upload thousands of annotated protein sequences as Multi-Fasta formatted files to run high-throughput BLAST searches against subject genomes. Similarly, the subject set also accepts newly sequenced genomes rapidly annotated via the xBASE Annotation server (10), and Multi-Fasta CDS and protein sets. A key innovation offered by mGenomeSubtractor is the ability to fragment genomes into thousands of short segments using a flexible sliding window algorithm, thus allowing for the identification of much shorter intergenic or intragenic unique sequences.
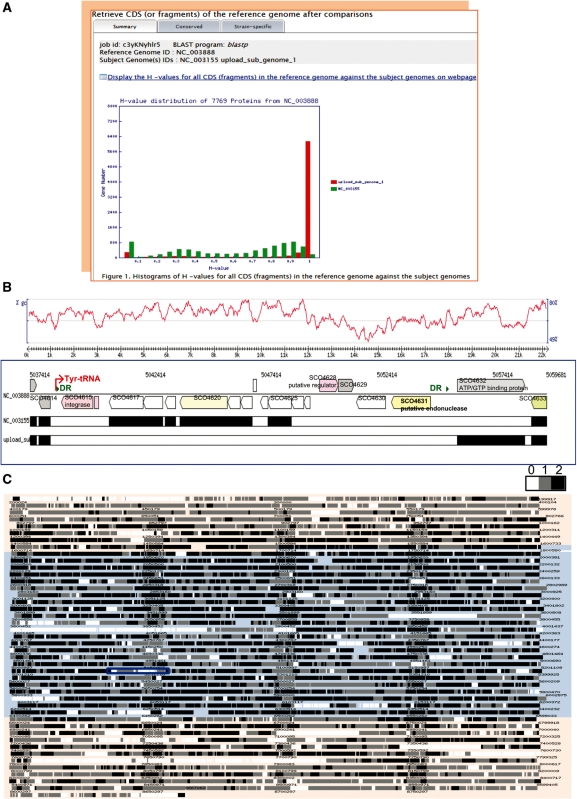
Figure 1.
In silico ‘subtractive hybridization’ based comparative genomics using mGenomeSubtractor with chromosomal sequences of Streptomyces coelicolor A3(2) (reference genome), S. avermitilis MA-4680 and S. lividans TK24 as the inputs. (A) Histogram of BLASTP-based Ha-values for all 7769 annotated chromosomal CDS in S. coelicolor A3(2) against all the annotated CDS in the two subject genomes, S. avermitilis MA-4680 (‘NC_003155’ in green) and S. lividans TK24 (‘upload_sub_genome_1’ in red). The Ha-value reflects the degree of similarity in terms of the length of match and the degree of identity at an amino acid level between the matching CDS in the subject genome and the query CDS examined. (B) An expanded view of the hypervariable region highlighted by a blue rectangle in (C) corresponding to the 17-kb SLP1-like island of S. coelicolor A3(2). The island boundaries are flanked with the Tyr tRNA gene (red arrow) and direct repeats (green triangles) as indicated. CDS are colour-coded based on their COG assignment. The S. coelicolor A3(2) unique SCO4231 codes for a putative Type IV restriction endonuclease that cleaves both Dcm methylated and Dnd phosphorothioated DNA. The two comparator genomes are shown below. Black bars indicate the extent of S. coelicolor A3(2) CDS within this region that are also present in the individual comparator genomes. A G+C profile of the selected S. coelicolor A3(2) region is shown topmost. (C) Chromosome map of S. coelicolor A3(2) with CDS colour-coded based on the number of comparator Streptomyces genomes identified as harbouring a amino acid sequence-conserved homologue. The core region spanning coordinates 1.5–6.4 Mb is highlighted with a light pink background while the 1.5 Mb left and 2.3 Mb right arms are backgrounded in sky blue (23). CDS shown in black (‘2’) are conserved across both the S. avermitilis MA-4680 and S. lividans TK24 comparator genomes, while at the other extreme those shown in white (‘0’) are unique to S. coelicolor A3(2). The strain-specific CDS were identified based on a Ha-value cutoff of <0.64.
Typically alignment is computed ‘on the fly’ in a matter of tens of seconds. Users are then directed to the retrieve page, which displays a histogram of H-values (Figure 1A) that helps users to set an appropriate cutoff to discriminate between conserved and strain-specific CDS. With the example shown, S. coelicolor A3(2) CDS were defined as conserved if the two H-values resulting from individual alignments versus S. lividans TK24 and S. avermitilis MA-4680 were >0.42. Advanced options such as the pre-definition of certain genomic regions as core or accessory and the option of matching to whole or partial subject set genomes allow users greater flexibility.
The Results page offers the following functional tabs for further analysis of the identified conserved or strain-specific CDS: ‘List’ [H-value, CDS annotation, multiple sequence alignment with MUSCLE (15) and Jalview (16), and/or primer design with Primer3Plus (17)], ‘Download’ (download sequence and annotation data in bulk), ‘Blast’ [BLAST search against the DEG (18), VFDB (19) and/or ACLAME (20) databases], ‘Core/Accessory Region’ (investigation of core and/or accessory genomic regions), ‘Design Probes’ [link to an integrated YODA (21) page for genome-wide CDS-specific probe selection for high-density oligonucleotide array construction]. mGenomeSubtractor also generates schematic outputs with zoom in utility if the reference genome is from the complete genome list or uploaded as a complete sequence with an associated annotation file in NCBI .ptt format. A colour-coded comparative genomic map of S. coelicolor A3(2) is shown in Figure 1C; CDS judged as shared using a Ha-value cutoff of 0.42 by none, either or both comparator genomes (S. lividans TK24 and S. avermitilis MA-4680) are as indicated. The 4776 S. coelicolor A3(2) CDS conserved across all three genomes shown in black with the legend label ‘2’ indicating conservation across both comparator genomes. Subsequent selection of the region of interest opens up a second and third graphical output comprising the G+C content and COG assignment of each CDS (Figures 1B and 2D).
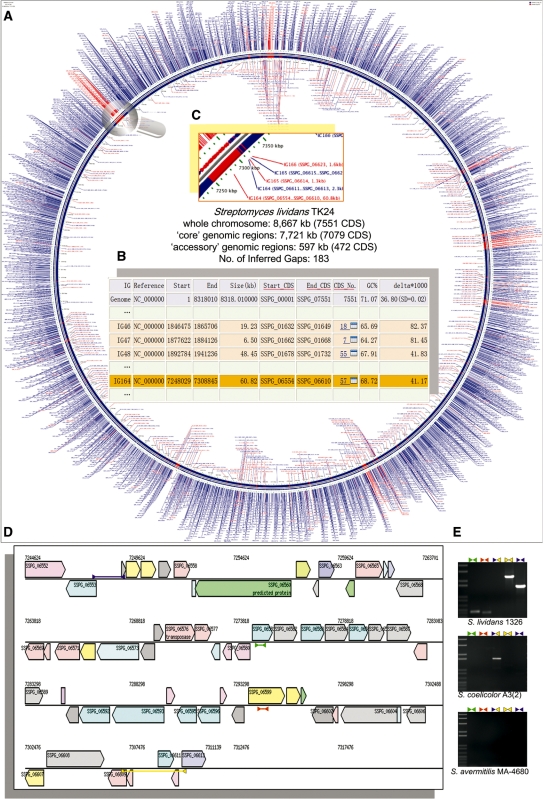
Figure 2.
In silico ‘subtractive hybridization’ of Streptomyces lividans TK24 against its close relative S. coelicolor A3(2) using mGenomeSubtractor. (A) Circular map of S. lividans TK24 linear chromosome showing inferred contigs (ICs) and inferred gaps (IGs). The ICs (in blue) are produced by merging adjacent genes classified as ‘conserved’ with the BLASTN-based H-values <0.81, whereas IGs (in red) comprise contiguous CDS defined as ‘strain-specific’. (B) Table containing feature details of mGenomeSubtractor-predicted S. lividans TK24 ‘accessory’ genomic regions borne on identified IGs. The variable region covering IG46, IG47 and IG48 highlighted in pink exhibits near identity to matching regions of the 94 kb dnd-encoding SLG island originally identified in S. lividans 1326. IG164 highlighted in orange is a likely novel S. lividans-specific island. (C) A magnified view of the IG164 island. (D) Schematic of IG164 with individual CDS color-coded by COG assignment. (E) PCR assays for detecting IG164 signals with the primers denoted as arrowheads in (D) targeting SSPG_06582 (green), SSPG_06599 (brown), and the left (purple) and right (yellow) IG164 boundaries. Primer3Plus (17) was integrated to facilitate design of PCR primers. DNA templates used were from S. lividans 1326 (top panel), S. coelicolor A3(2) (middle) and S. avermitilis MA-4680 (bottom), respectively.
The Results page ‘Core/Accessory Region’ tab outputs mGenomeSubtractor-generated ‘core’ and/or ‘accessory’ reference genome regions by merging adjacent genes classified as ‘conserved’ or ‘strain-specific’. Accessory regions are tabulated to detail boundary coordinates and classical features of horizontally acquired genomic islands such as immediate flanking tRNA genes, internal integrase and/or transposase genes, and genome-atypical G+C content and/or dinucleotide distribution bias. The mGenomeSubtractor web-server also generates a graphical output comprising a circular map of the reference genome that highlights the locations and sizes of identified core and accessory regions within this reference template (Figure 2A).
mGenomeSubtractor::Gene Pool for estimate of species-specific gene pool
The size of a species gene pool, comprising the complete set of unique genes in a particular species, is probably a function of the extent of genetic diversity within and close to the evolutionary branch bearing the aforementioned species. The mpiBLAST-based mGenome-Subtractor::Gene Pool tool allows for estimation and classification of the known species-specific gene pool as represented within the set of sequenced genomes analysed. Supplementary Figure S3 shows the mGenome-Subtractor::Gene Pool Input and Results pages corresponding to the example analysis of seven completely sequenced Pseudomonas aeruginosa chromosomes as the input, four NCBI complete genomes (LESB58, PA7, UCBPP-PA14 and PAO1) and other three sequences (PACS2, C3719 and 2192) annotated by xBASE server (10). Firstly, all 5925 annotated CDS present on the P. aeruginosa LESB58 genome were included in the gene pool. Then each annotated CDS on the P. aeruginosa PA7 genome was used as a query in an mpiBLASTN search against the LESB58 genome sequence. The 1003 PA7-specific CDS identified by mGenomeSubtractor::Gene Pool (H-value <0.42) based on this two genome comparison were added to the gene pool. Next, a similar reiterative process was used to analyse each of the remaining five P. aeruginosa genomes leading to the sequential inclusion of a further 460 genes into the growing gene pool. Finally, single representative unique CDSs were selected from the gene pool by removing redundant ‘duplicated’ genes as determined using the H-value criteria (H-value >0.81). mGenome-Subtractor::Gene Pool analysis of the seven complete P. aeruginosa genomes using the criteria stated above defined a known P. aeruginosa gene pool comprising 7285 unique CDS, a number that exceeded the total number of non-duplicated CDS in any one chromosome by 16–39%.
The Results page displays summary data of the stored gene sequences, associated annotation information and COG classification categories as interactive tables, bar charts and pie charts (Supplementary Figure S3C–H). Users can also select ‘Design oligonucleotide probes with YODA’ to aid production of high-density oligonucleotide arrays representing entire defined gene pools or particular sub-pools of interest. YODA (21) can be run with default parameters or with user-defined constraints.
IMPLEMENTATION
The mGenomeSubtractor server consists of two basic components: the web interface and the computational pipeline. The computational component is written in Perl/Bioperl and uses mpiBLAST as the core module for checking nucleotide/amino acid sequence similarity. mpiBLAST (http://www.mpiblast.org/), a parallel implementation of NCBI BLAST using the message passing interface (MPI) library, allows time-consuming local alignment processes to be done in parallel on computing cluster architecture. The following freely available components were integrated by the mGenomeSubtractor server: (i) oligo/primer design tools, YODA (21) and Primer3Plus (17); (ii) multiple sequence alignment and visualization tools (MUSCLE) (15) and Jalview (16); (iii) genome visualization tool CGview (22); (iv) database of essential genes (DEG) (18); (v) a classification of genetic mobile elements (ACLAME) (20); (vi) virulence factors database (VFDB) (19). mGenomeSubtractor is now run on a high-performance 23-node cluster (21 computing nodes, one management node and one storage node) interconnected with a 1000 Mb ethernet network. Each computing node is equipped with two quad-core Xeon 2.33 GHz processors and 8 GB of RAM.
Compared to our previously reported stand-alone tool GenomeSubtractor 1.0 (11,12), mpiBLAST-facilitated mGenomeSubtractor offers the following major enhancements: (i) several orders of magnitude accelerated sequence alignment by up to 168 cores; (ii) query genome fragmentation using a flexible sliding window algorithm; (iii) facilitation of preliminary functional analyses of core/accessory genomic regions supported by powerful interactive schematic outputs and tabulated data of sequence/CDS features; (iv) automated extraction, assembly and summary analyses of species-specific gene pools based on input genome sets. Presently, mGenome-Subtractor takes ∼1 min to compare all 5312 CDS in the 5.5 Mb Escherichia coli O157:H7 EDL933 chromosome against 23 completely sequenced divergent E. coli chromosomes. In addition, mGenomeSubtractor displays a URL for subsequent retrieval of results for jobs demanding exceedingly large amounts of computing and hence requiring greater than a minute for completion. Alternatively, results can be emailed automatically upon completion of the job. Each run is assigned a job-id and associated output files are stored on the server for 7 days.
APPLICATIONS
See Figures 1 and 2, and Supplementary Figure S3, and more details in Supplementary Data for examples of applications of mGenomeSubstractor.
CONCLUSION
The mGenomeSubtractor web server, which integrates a wide variety of useful analytical and functional tools, has been developed to perform mpiBLAST-based single-process comparison of a reference bacterial genome against up to 30 user-selected genomes and/or 10 user-supplied genomes. With parallel computing PC cluster architecture, ease of navigation and flexible input options, we present it as a quick and comprehensive genome-mining tool dedicated to biologists. In a near future version of mGenomeSubtractor, we will improve its computing power markedly with enhanced inter-node communication over a 20 Gb Infiniband network. The upgraded version will be able to directly align mass short-read sequence data against selected bacterial genomes or even the full catalogue of finished microbial genomes, thus providing an invaluable analytical tool to help exploit the next generation sequencing data ‘tsunami’ in our midst. We propose that a tool such as mGenomeSubtractor will support the rapidly escalating number of comparative bacterial genomics studies aimed at defining genomic biomarkers of evolutionary lineage, phenotype, pathotype, environmental adaptation and/or disease-association of diverse bacterial species.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Natural Science Foundation of China (30871345, 30700013, 30970080 and 30821005); a Royal Society—National Natural Science Foundation of China International Joint Project grant (2007/R3 to to K.R. and Z.D.); Chen Xing young scholars programme, Shanghai Jiaotong University (to H.Y.O.). Funding for open access charge: National Natural Science Foundation of China.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful to Prof. Chun-Ting Zhang and Dr Ren Zhang for their supporting work. We thank the Broad Institute of MIT and Harvard and the Genome Center at Washington University for their policy of making preliminary sequence data publicly available and acknowledge the use in this study of unpublished genome data of S. lividans strain TK24, P. aeruginosa strain C3719, 2192 and PACS2.
REFERENCES
- 1.Medini D, Donati C, Tettelin H, Masignani V, Rappuoli R. The microbial pan-genome. Curr. Opin. Genet. Develop. 2005;15:589–594. doi: 10.1016/j.gde.2005.09.006. [DOI] [PubMed] [Google Scholar]
- 2.Binnewies TT, Motro Y, Hallin PF, Lund O, Dunn D, La T, Hampson DJ, Bellgard M, Wassenaar TM, Ussery DW. Ten years of bacterial genome sequencing: comparative-genomics-based discoveries. Funct. Integr. Genomics. 2006;6:165–185. doi: 10.1007/s10142-006-0027-2. [DOI] [PubMed] [Google Scholar]
- 3.Ou HY, Smith R, Lucchini S, Hinton J, Chaudhuri RR, Pallen M, Barer MR, Rajakumar K. ArrayOme: a program for estimating the sizes of microarray-visualized bacterial genomes. Nucleic Acids Res. 2005;33:e3. doi: 10.1093/nar/gni005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hacker J, Blum-Oehler G, Muhldorfer I, Tschape H. Pathogenicity islands of virulent bacteria: structure, function and impact on microbial evolution. Mol. Microbiol. 1997;23:1089–1097. doi: 10.1046/j.1365-2958.1997.3101672.x. [DOI] [PubMed] [Google Scholar]
- 5.Frost LS, Leplae R, Summers AO, Toussaint A. Mobile genetic elements: the agents of open source evolution. Nature Rev. Microbiol. 2005;3:722–732. doi: 10.1038/nrmicro1235. [DOI] [PubMed] [Google Scholar]
- 6.Abbott JC, Aanensen DM, Rutherford K, Butcher S, Spratt BG. WebACT–an online companion for the Artemis Comparison Tool. Bioinformatics. 2005;21:3665–3666. doi: 10.1093/bioinformatics/bti601. [DOI] [PubMed] [Google Scholar]
- 7.Grant JR, Stothard P. The CGView Server: a comparative genomics tool for circular genomes. Nucleic Acids Res. 2008;36:W181–W184. doi: 10.1093/nar/gkn179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kurtz S, Phillippy A, Delcher AL, Smoot M, Shumway M, Antonescu C, Salzberg SL. Versatile and open software for comparing large genomes. Genome Biol. 2004;5:R12. doi: 10.1186/gb-2004-5-2-r12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Darling AC, Mau B, Blattner FR, Perna NT. Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 2004;14:1394–1403. doi: 10.1101/gr.2289704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chaudhuri RR, Loman NJ, Snyder LA, Bailey CM, Stekel DJ, Pallen MJ. xBASE2: a comprehensive resource for comparative bacterial genomics. Nucleic Acids Res. 2008;36:D543–D546. doi: 10.1093/nar/gkm928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ou HY, Chen LL, Lonnen J, Chaudhuri RR, Thani AB, Smith R, Garton NJ, Hinton J, Pallen M, Barer MR, et al. A novel strategy for the identification of genomic islands by comparative analysis of the contents and contexts of tRNA sites in closely related bacteria. Nucleic Acids Res. 2006;34:e3. doi: 10.1093/nar/gnj005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ou HY, He X, Harrison EM, Kulasekara BR, Thani AB, Kadioglu A, Lory S, Hinton JC, Barer MR, Deng Z, et al. MobilomeFINDER: web-based tools for in silico and experimental discovery of bacterial genomic islands. Nucleic Acids Res. 2007;35:W97–W104. doi: 10.1093/nar/gkm380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ou HY, Ju CT, Thong KL, Ahmad N, Deng Z, Barer MR, Rajakumar K. Translational genomics to develop a Salmonella enterica serovar Paratyphi A multiplex polymerase chain reaction assay. J. Mol. Diagnostics. 2007;9:624–630. doi: 10.2353/jmoldx.2007.070064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fukiya S, Mizoguchi H, Tobe T, Mori H. Extensive genomic diversity in pathogenic Escherichia coli and Shigella Strains revealed by comparative genomic hybridization microarray. J. Bacteriol. 2004;186:3911–3921. doi: 10.1128/JB.186.12.3911-3921.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Waterhouse AM, Procter JB, Martin DM, Clamp M, Barton GJ. Jalview Version 2–a multiple sequence alignment editor and analysis workbench. Bioinformatics. 2009;25:1189–1191. doi: 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Untergasser A, Nijveen H, Rao X, Bisseling T, Geurts R, Leunissen JA. Primer3Plus, an enhanced web interface to Primer3. Nucleic Acids Res. 2007;35:W71–W74. doi: 10.1093/nar/gkm306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang R, Lin Y. DEG 5.0, a database of essential genes in both prokaryotes and eukaryotes. Nucleic Acids Res. 2009;37:D455–D458. doi: 10.1093/nar/gkn858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang J, Chen L, Sun L, Yu J, Jin Q. VFDB 2008 release: an enhanced web-based resource for comparative pathogenomics. Nucleic Acids Res. 2008;36:D539–D542. doi: 10.1093/nar/gkm951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Leplae R, Lima-Mendez G, Toussaint A. ACLAME: a CLAssification of mobile genetic elements, update 2010. Nucleic Acids Res. 2010;38:D57–D61. doi: 10.1093/nar/gkp938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nordberg EK. YODA: selecting signature oligonucleotides. Bioinformatics. 2005;21:1365–1370. doi: 10.1093/bioinformatics/bti182. [DOI] [PubMed] [Google Scholar]
- 22.Stothard P, Wishart DS. Circular genome visualization and exploration using CGView. Bioinformatics. 2005;21:537–539. doi: 10.1093/bioinformatics/bti054. [DOI] [PubMed] [Google Scholar]
- 23.Bentley SD, Chater KF, Cerdeno-Tarraga AM, Challis GL, Thomson NR, James KD, Harris DE, Quail MA, Kieser H, Harper D, et al. Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2) Nature. 2002;417:141–147. doi: 10.1038/417141a. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.