Abstract
G-language genome analysis environment (G-language GAE) contains more than 100 programs that focus on the analysis of bacterial genomes, including programs for the identification of binding sites by means of information theory, analysis of nucleotide composition bias and the distribution of particular oligonucleotides, calculation of codon bias and prediction of expression levels, and visualization of genomic information. We have provided a collection of web services for these programs by utilizing REST and SOAP technologies. The REST interface, available at http://rest.g-language.org/, provides access to all 145 functions of the G-language GAE. These functions can be accessed from other online resources. All analysis functions are represented by unique universal resource identifiers. Users can access the functions directly via the corresponding universe resource locators (URLs), and biological web sites can readily embed the functions by simply linking to these URLs. The SOAP services, available at http://www.g-language.org/wiki/soap/, provide language-independent programmatic access to 77 analysis programs. The SOAP service Web Services Definition Language file can be readily loaded into graphical clients such as the Taverna workbench to integrate the programs with other services and workflows.
INTRODUCTION
When conducting a scientific research project, a considerable amount of time and effort is often expended in the process of designing effective protocols or methodology. This process comprises a larger proportion of the work load in hypothesis-free or hypothesis-generating disciplines such as bioinformatics and computational genomics (1), which generally involve knowledge discovery and data mining based on heuristic approaches. Research in these areas often requires the comparative testing of numerous software tools with the use of hundreds of parameters, and a multitude of input sequences from a variety of data sources. The G-language genome analysis environment (G-language GAE; http://www.g-language.org/) (2,3) has been developed as a workbench to expedite routine heuristic research processes in computational genomics. It provides a set of tools, software libraries and interfaces for use primarily in bacterial genome analysis. Because the G-language GAE is comprised of a set of Perl libraries, it is compatible with BioPerl, but it is 7–32 times faster than BioPerl at parsing GenBank files (see http://www.g-language.org/wiki/benchmark for benchmarking details), and it has a more intuitive gateway interface. For example, the G-language GAE can parse and load sequence data from local flat files in numerous file formats, or from remote databases by using accession numbers, or from data objects created with BioPerl (4), all with a single gateway function with automatic interpretation of the data location and type. Moreover, G-language GAE is equipped with an interactive Perl/UNIX shell, which supports a persistent workspace, so a user does not need to write or edit a script in every trial-and-error routine, but instead can interactively test one method at a time. The software package also contains more than 100 original analysis programs, including those for the identification of binding sites with the use of information theory, analysis of nucleotide composition bias, analysis of the distribution of characteristic oligonucleotides, analysis of codons and prediction of expression levels, and visualization of genomic information [for a comprehensive review describing the detailed algorithms for each of these tools, see ref. (5)]. Some of the analysis capabilities are trivial, such as the tools for global alignment and for basic statistics, and for some of the programs similar functionality is available in comprehensive suites like EMBOSS (6). On the other hand, the G-language GAE contains a more comprehensive collection of tools for codon analysis than that found in most popular packages, such as CodonW (http://codonw.sourceforge.net/), and a number of published tools are only available in this system: for example, accurate prediction of the origin and terminus of replication in bacteria using noise-reduction filtering with the fast fourier transform algorithm (7), quantification of replication-related mutation or selection bias on bacterial chromosomes and plasmids (8,9), measurement of codon bias by using the weighted sum of relative entropy (10), accurate detection of the long-term host of a plasmid by measuring the Mahalanobis distance of the genomic signatures (11), interactive and zoomable chaos game representation (12), and several visualization tools for genomic information using the google maps application programming interface (API) (13).
With the availability of thousands of software tools and an ever increasing wealth of genomic resources, interoperability has become an essential requirement for bioinformatics tools. The availability of tools as web services provides interoperability, as well as other advantages such as a lack of cost for installation and maintenance (14). The two major web service technologies that are widely adopted in the bioinformatics community are SOAP (http://www.w3.org/TR/soap/) with web services interoperability (WS-I; http://www.ws-i.org) guidelines and representational state transfer (REST; http://www.ics.uci.edu/∼fielding/pubs/dissertation/rest_arch_style.htm). Each has complementing advantages (15). SOAP is a traditional web service protocol that exchanges structured information using XML-based messages. SOAP requires the use of dedicated software libraries, and hence the messages are not meant to be human-readable. The protocol is suited for programmatic access with defined data types. Numerous bioinformatics services are already available as SOAP-based web services, and a number of graphical client software packages for the construction of workflows utilize these services: for example, the Taverna workbench (16). In contrast, REST is laissez faire approach without strict definitions for its protocol, that is recently revived in light of its advantages over SOAP. Basically, REST provides universal resource locator (URL)-based access over hypertext transfer protocol (HTTP), and therefore can be easily utilized with any tools that can access the web, such as web browsers. Moreover, each REST operation has a unique universal resource identifier (URI), which can be used to represent the service as an online resource that can be utilized from other web sites as a hyperlink. In short, SOAP is suited for operation-centric access by programming languages, and REST is suited for quick resource-centric access without the requirement of specialized tools. Therefore, we provide a total of 145 services encompassing 21 categories (Table 1), in SOAP and/or REST interfaces. In the following sections, we describe the system architecture and provide usage examples of these services.
Table 1.
Available services
| Service class (number of Web APIs) | Description |
|---|---|
| G (39), Primitive (7), Utils (6) | Basic sequence manipulation and data access |
| Alignment (2) | Sequence alignment |
| AminoAcid (3) | Calculation of properties of amino acids |
| Codon (19) | Calculation of codon usage and prediction of expression levels |
| Consensus (6) | Prediction of sequence motifs using molecular information theory |
| Gcskew (13) | Analysis of skews in genome composition, prediction of replication origin and terminus |
| GenomeMap (7), Graph/Gmap (2) | Visualization of genomic information |
| OverLapping (1) | Identification of overlapping genes |
| PatSearch (15) | Analysis of oligomer frequencies, calculation of genomic signatures |
| Help (2) | Documentation |
| Statistics (13) | Basic statistics |
| WebServices (3), Eutils (2), BLAST (1), EMBOSS (1), COG (1), Operon (1), PEC (1) | Simplified interface to external web services |
| Total (145) |
REST SERVICES
REST services are suited for resource-centric access and are extremely easy to use without the need for programming or specialized tools. We therefore provide basic data parsing functionality as well as all 145 functions of the G-language GAE in our set of REST services. While the use of XML as an exchange format is sometimes recommended even for REST services, all of our services return the results in plain text for human-readability and simplicity, and status and error reports comply with the HTTP status code: 200 for successful request, 404 for invalid request and 302 for URL transfer to result image files. Service operations are effectively cached on the server for optimized performance. Each service operation is represented by two unique URLs following a simple syntax, with the base URL being either http://rest.g-language.org/ or http://useG.jp/. These two URLs point to an identical server, and in the following descriptions we use the latter.
An overview of the URL syntax is presented in Figure 1. Parsing a flat file containing a DNA sequence requires three input fields: [sequence], [gene] and [feature name] (Figure 1A). For example, in order to obtain the start position of the recA gene in the Escherichia coli K-12 MG1655 genome (accession number refseq:NC_000913 or embl:U00096), a user can access the corresponding unique URL http://useG.jp/NC_000913/recA/start to obtain the position ‘2 820 730’. The first field [sequence] can be (i) an accession number of one of the GenBank files available on our server (1247 files containing complete genome and plasmid sequences, at the time of writing; see http://useG.jp/organism_list/ for a comprehensive listing), (ii) built-in genomes in the G-language GAE, such as ecoli, mgen, bsub, cyano, pyro, corresponding to E. coli, Mycoplasma genitalium, Bacillus subtilis, Synechococcus sp. and Pyrococcus furiosus, respectively, (iii) a uniform sequence address (USA) in the form of [database name]:[accession number] such as ‘refseq:NC_000913’ to automatically retrieve a DNA or protein sequence from UniProt (swiss), GenBank (genbank), GenPept (genpept), EMBL (embl) and RefSeq (refseq), or (iv) a temporary six digit hexadecimal ID given to an imported sequence that is in a format supported by BioPerl, such as GenBank, EMBL, FastA, FastQ and SwissProt, and has been uploaded using the function at http://useG.jp/upload/. Therefore, input of ‘NC_000913’, ‘ecoli’, ‘refseq:NC_000913’, ‘embl:U00096’, or the temporary six-digit ID in the field [sequence] leads to identical results. The second input field [gene] can be the common gene symbol (e.g. recA), the canonical gene ID (e.g. b2699), or the feature ID in the G-language GAE (e.g. FEATURE5804 or CDS2646). To obtain multiple entries of interest, the [sequence] input can also be a wild-card (*) or a search field-value pair; for example, to search for the keyword ‘metabolism’ in the ‘product’ feature, the [sequence] input is ‘product = metabolism’). When multiple genes are specified in this way, results are given in multi-FASTA format with gene names as sequence IDs. The last field [feature name] is the name of the feature key in genome flat files (such as start, end, direction, translation, db_xref).
Figure 1.
Syntax for G-language REST service.
Using similar syntax, users can easily utilize the 145 analysis programs of the G-language GAE by specifying the program name in the field [method], and by appending optional parameters when necessary in the field [option = value] (Figure 1B and C). For example, the retrieval of all the 100-bp nt sequences that are immediately upstream of the start codon of genes in E. coli, in a multi-FASTA format, can be achieved using http://useG.jp/NC_000913/*/before_startcodon, GC skew analysis can be performed using http://useG.jp/NC_000913/gcskew/, and a cumulative GC skew with a 1 kb window can be calculated using http://useG.jp/NC_000913/gcskew/cumulative = 1/window = 1000/. The genome sequence analysis programs often return the results in graphical form, so that the user can readily interpret the results (Figure 2).
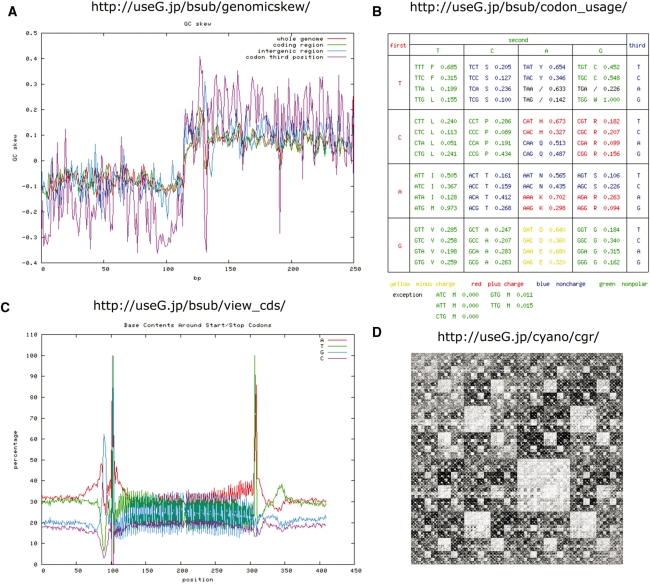
Figure 2.
Examples of graphical analysis results. (A) GC skew graph calculated in all regions of the entire genome (red), coding regions (green), intergenic regions (blue) and third nucleotide in codons (purple) with genomicskew function in B. subtilis genome. (B) Codon table of B. subtilis genome visualized with codon_usage function. (C) Nucleotide frequency in regions of 100 bp around start and stop codons using all genes within the genome of B. subtilis using view_cds function. Strong conservation of nucleotides at start and stop codons, as well as conserved upstream Shine–Dalgarno sequence can be observed. (D) Chaos game representation visualized for Synechococus sp.
Here we have implemented our REST web service to directly return result text and images in arbitrary text and URLs, mostly for intuitive usage by biologists from web browsers. On the other hand, RESTful resources should ideally be marked up with resource URIs for semantic interoperability (17), which will be our future work.
SOAP SERVICES
The G-language SOAP services include 77 operation-centric analyses, and exclude simple data access and manipulation services that are better handled with REST. All services return the results synchronously without polling, and take either single scalar values (64 methods) or lists (13 methods) as their main input (named according to the input type), followed by a hash of optional parameters (named ‘params’), as demonstrated in the following Perl program. As is the case for REST services, the input sequence can be a RefSeq or G-language GAE ID, a USA of remote data, or a raw DNA or protein sequence in a file format supported in BioPerl. The types of output values are scalar, list, or a URL of the result file. These services are based on SOAP 1.1, and service descriptions are listed in Web Services Definition Language (WSDL) 1.1 at http://soap.g-language.org/g-language.wsdl. The following is an example of a Perl program that utilizes our SOAP service to calculate and visualize cumulative GC skew in the E. coli genome; it accesses the ‘gcskew’ service with the ‘-cumulative’ option set to 1.
#!/usr/bin/env perl
use SOAP::Lite;
$soap = SOAP::Lite->service(“http://soap.g-language.org/g-language.wsdl”);
#set parameters
$in0 = SOAP::Data->new(name=>'in0', value=>’ecoli’);
%param = (-cumulative=>1);
$inputParams = SOAP::Data->name(‘params')->type(map=>\%param);
#run the web service “gcskew”
print $soap->gcskew($in0, $inputParams);
One of the primary advantages of SOAP services is their ability to integrate with hundreds of other bioinformatics services that are already available with SOAP technology, in the form of workflows. Because the WSDL file for our SOAP service includes the description of all 77 services within one file, users can readily incorporate all these services into the Taverna workbench by loading this WSDL file. Moreover, nine example workflows utilizing G-language SOAP services have been submitted to the myExperiment web site (http://www.myexperiment.org/) (18,19) that can be loaded in Taverna, and users can download and customize these workflows according to their specific needs (http://www.myexperiment.org/search?query=g-language&type=all). Figure 3 shows one of the workflows loaded in Taverna, namely the ‘bacteria analysis system’ (ID:779 in myExperiment), which runs 12 analysis services based on a given bacterial genome.
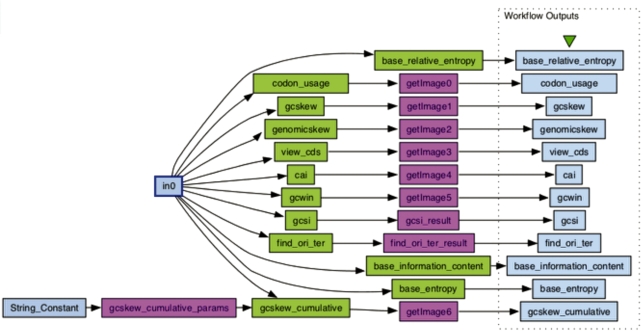
Figure 3.
Example workflow loaded in Taverna. This workflow calculates and graphs the DNA sequence conservation around the start codon in terms of relative entropy (base_relative_entropy), entropy (base_entropy) and information content (base_information_content) (20), calculates the codon usage (codon_usage), graphs the GC skew (gcskew), cumulative GC skew (gcskew_cumulative), GC skew in multiple regions of genome (genomicskew) (21), visualizes the nucleotide composition around the coding regions (view_cds), calculates the Codon Adaptation Index and predicts gene expression levels (cai) (22), visualizes the GC content (gcwin), calculates the GC Skew Index (gcsi) (8), and predicts the origin and terminus of replication (find_ori_ter) (7).
APPLICATIONS
The Perl API for the G-language GAE provides powerful features that aid programmatic sequence manipulation; however, installation of the API needs expert knowledge because the API requires many external modules. Therefore, a more lightweight version of the G-language GAE API has been developed as a wrapper around the REST services. This module, named Bio::Glite, is available from the comprehensive perl archive network (CPAN; http://www.cpan.org/), and can be easily installed using the command ‘cpan Bio::Glite’ in any UNIX systems with Perl installed. Users of this module can utilize the G-language REST services with the same usability as the original Perl API.
DOCUMENTATION
Complete documentation about each methods and services are available at AJAX document center (http://ws.g-language.org/gdoc/). A list of all 145 G-language REST services is available at http://useG.jp/method_list/gb/ and http://useG.jp/method_list/ mainly for programmatic access, and detailed documentation about each of the services, including service description, example usage, optional parameters and their default values can be viewed at http://useG.jp/help/[method], where [method] is the input field for the function name. Further documentation and usage examples are available at http://rest.g-language.org/. For SOAP services, example scripts in Perl, Ruby and Python languages and detailed documentation, are available at http://www.g-language.org/wiki/soap/. The SOAP services have also been registered in the BioCatalogue(http://www.biocatalogue.org/services/2623-glangsoapservice_651637).
CONCLUSIONS
We have designed and implemented a set of web service interfaces to the G-language GAE, which allow access to 145 analysis programs through REST and SOAP web service technologies. The REST services enable quick and easy access to all programs from any web browser through unique URLs, and thus the services can be accessed, in the form of hyperlinks, from other online resources. The generic handling of sequence data, with automatic interpretation of multiple identifiers and file formats, allows easy linking to a wide variety of resources. The 77 SOAP services enable programmatic access to the G-language GAE by programs written in languages other than Perl, such as Ruby, Python and Java. By using workflow management software such as Taverna, all G-language SOAP services can be readily integrated with other bioinformatics web services that have rich graphical user interfaces.
FUNDING
Grant-in-Aid for Young Scientists No. 20710158 from the Japan Society for the Promotion of Science (JSPS), and by funds from the Yamagata Prefectural Government and Tsuruoka City. Funding for open access charge: Funds from the Yamagata Prefectural Government and Tsuruoka City.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The development of the web services was significantly facilitated by the DBCLS BioHackathon 2009, jointly hosted by the Database Center for Life Science (DBCLS) and Okinawa Institute of Science and Technology (OIST). We thank the members of G-language Project at the Institute for Advanced Biosciences, Keio University, and elsewhere, for their extremely valuable feedback and support.
REFERENCES
- 1.Kell DB, Oliver SG. Here is the evidence, now what is the hypothesis? The complementary roles of inductive and hypothesis-driven science in the post-genomic era. Bioessays. 2004;26:99–105. doi: 10.1002/bies.10385. [DOI] [PubMed] [Google Scholar]
- 2.Arakawa K, Mori K, Ikeda K, Matsuzaki T, Kobayashi Y, Tomita M. G-language Genome Analysis Environment: a workbench for nucleotide sequence data mining. Bioinformatics. 2003;19:305–306. doi: 10.1093/bioinformatics/19.2.305. [DOI] [PubMed] [Google Scholar]
- 3.Arakawa K, Tomita M. G-language System as a platform for large-scale analysis of high-throughput omics data. J. Pesticide Sci. 2006;31:282–288. [Google Scholar]
- 4.Stajich JE, Block D, Boulez K, Brenner SE, Chervitz SA, Dagdigian C, Fuellen G, Gilbert JG, Korf I, Lapp H, et al. The Bioperl toolkit: Perl modules for the life sciences. Genome Res. 2002;12:1611–1618. doi: 10.1101/gr.361602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Arakawa K, Suzuki H, Tomita M. Computational genome analysis using the G-language system. Genes Genomes Genomics. 2008;2:1–13. [Google Scholar]
- 6.Rice P, Longden I, Bleasby A. EMBOSS: the European molecular biology open software suite. Trends Genet. 2000;16:276–277. doi: 10.1016/s0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]
- 7.Arakawa K, Saito R, Tomita M. Noise-reduction filtering for accurate detection of replication termini in bacterial genomes. FEBS Lett. 2007;581:253–258. doi: 10.1016/j.febslet.2006.12.021. [DOI] [PubMed] [Google Scholar]
- 8.Arakawa K, Suzuki H, Tomita M. Quantitative analysis of replication-related mutation and selection pressures in bacterial chromosomes and plasmids using generalised GC skew index. BMC Genomics. 2009;10:640. doi: 10.1186/1471-2164-10-640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Arakawa K, Tomita M. The GC skew index: a measure of genomic compositional asymmetry and the degree of replicational selection. Evol. Bioinform. Online. 2007;3:159–168. [PMC free article] [PubMed] [Google Scholar]
- 10.Suzuki H, Saito R, Tomita M. The ‘weighted sum of relative entropy': a new index for synonymous codon usage bias. Gene. 2004;335:19–23. doi: 10.1016/j.gene.2004.03.001. [DOI] [PubMed] [Google Scholar]
- 11.Suzuki H, Sota M, Brown CJ, Top EM. Using Mahalanobis distance to compare genomic signatures between bacterial plasmids and chromosomes. Nucleic Acids Res. 2008;36:e147. doi: 10.1093/nar/gkn753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Arakawa K, Oshita K, Tomita M. A web server for interactive and zoomable Chaos game representation images. Source Code Biol. Med. 2009;4:6. doi: 10.1186/1751-0473-4-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Arakawa K, Tamaki S, Kono N, Kido N, Ikegami K, Ogawa R, Tomita M. Genome Projector: zoomable genome map with multiple views. BMC Bioinformatics. 2009;10:31. doi: 10.1186/1471-2105-10-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stein L. Creating a bioinformatics nation. Nature. 2002;417:119–120. doi: 10.1038/417119a. [DOI] [PubMed] [Google Scholar]
- 15.Stockinger H, Attwood T, Chohan SN, Cote R, Cudre-Mauroux P, Falquet L, Fernandes P, Finn RD, Hupponen T, Korpelainen E, et al. Experience using web services for biological sequence analysis. Brief Bioinform. 2008;9:493–505. doi: 10.1093/bib/bbn029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hull D, Wolstencroft K, Stevens R, Goble C, Pocock MR, Li P, Oinn T. Taverna: a tool for building and running workflows of services. Nucleic Acids Res. 2006;34:W729–W732. doi: 10.1093/nar/gkl320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Richardson L, Ruby S. RESTful Web Services. Tokyo: O'Reilly Media; 2007. [Google Scholar]
- 18.De Roure D, Goble C, Bhagat J, Cruickshank D, Goderis A, Michaelides D, Newman D. In eScience '08. IEEE Fourth International Conference. Los Alamos: IEEE Computer Society; 2008. myExperiment: Defining the Social Virtual Research Environment; pp. 182–189. [Google Scholar]
- 19.De Roure D, Goble C, Stevens R. The design and realisation of the Virtual Research Environment for social sharing of workflows. Future Gener. Comp. Syst. 2009;25:561–567. [Google Scholar]
- 20.Schneider TD. Consensus sequence Zen. Appl Bioinformatics. 2002;1:111–119. [PMC free article] [PubMed] [Google Scholar]
- 21.Lobry JR. Asymmetric substitution patterns in the two DNA strands of bacteria. Mol. Biol. Evol. 1996;13:660–665. doi: 10.1093/oxfordjournals.molbev.a025626. [DOI] [PubMed] [Google Scholar]
- 22.Sharp PM, Li WH. The codon Adaptation Index—a measure of directional synonymous codon usage bias, and its potential applications. Nucleic Acids Res. 1987;15:1281–1295. doi: 10.1093/nar/15.3.1281. [DOI] [PMC free article] [PubMed] [Google Scholar]