Abstract
The EMBRACE (European Model for Bioinformatics Research and Community Education) web service collection is the culmination of a 5-year project that set out to investigate issues involved in developing and deploying web services for use in the life sciences. The project concluded that in order for web services to achieve widespread adoption, standards must be defined for the choice of web service technology, for semantically annotating both service function and the data exchanged, and a mechanism for discovering services must be provided. Building on this, the project developed: EDAM, an ontology for describing life science web services; BioXSD, a schema for exchanging data between services; and a centralized registry (http://www.embraceregistry.net) that collects together around 1000 services developed by the consortium partners. This article presents the current status of the collection and its associated recommendations and standards definitions.
INTRODUCTION
Since the early days of the web, the life science community has embraced its use as a mechanism for sharing data, software and knowledge. The enthusiasm and willingness to exchange both research results and the tools necessary to access, visualize and analyse those data is evident through the ever-growing number of resources reported in the annual web server (1) and database editions (2) of Nucleic Acids Research (NAR). More recently, the need has been recognised to provide not only human-accessible web pages but also programmatic access to the same resources via so-called ‘Web services’ (3–6). In terms of experimental scalability and reproducibility, there are obvious advantages to being able to automate access to these remote resources through the use of programming languages or workflow systems, such as Taverna (7) and Kepler (8). However, the use of web services is not without its problems. Summarizing Hull et al. (7), these have historically included: (i) reliance on complex and evolving underlying technologies prone to generating cryptic error messages, (ii) limited documentation and metadata describing services; (iii) incompatible and inconsistent inputs and outputs between services; and (iv) unpredictable performance/reliability. In an effort to address these problems and to provide the life science community with a collection of coherent and robust bioinformatics web services, in 2005, the European Commission provided funds to establish the EMBRACE (European Model for Bioinformatics Research and Community Education) Network of Excellence. The consortium, consisting of 18 institutions, brought together providers of major tools and databases with experts from the informatics domain. To date, the project has produced almost 1000 services, covering a wide functional spectrum, from traditional programs, such as BLAST (9) and ClustalW (10), through to more domain-specific tools and resources, such as metabolite substructure prediction from GC–MS profiles (11) or the prediction of protein stabilization by introducing prolines into the structure (12).
EMBRACE has developed technologies and recommendations to improve the use and uptake of web services within the life science domain; the relationship of these solutions to the issues identified by Hull et al. (7) are outlined in Table 1.
Table 1.
The problems identified in Hull et al. (7) and their corresponding solutions, as developed by the EMBRACE consortium
| Problem | EMBRACE solution |
|---|---|
| Inconsistent use of technology | EMBRACE Technology Recommendation Documents. Provide guidance for producers and consumers on selecting appropriate technologies and standards for developing life science web services. |
| Limited service metadata | EDAM Ontology. A vocabulary of terms and relations suitable for annotating the behaviour, inputs and outputs of life science services, and for associating meaning with data exchanged between services. |
| Incompatible interfaces | BioXSD data interchange format definition. A mechanism for unifying the exchange of data between bioinformatics services. |
| Unreliable services | EMBRACE Active Registry. A mechanism for finding services and for monitoring their performance. |
SELECTING A SUITABLE WEB SERVICE TECHNOLOGY
Establishing a basic form of technological standardization required the consortium to first agree on a consistent definition of ‘Web service’. The original term was defined by the World Wide Web Consortium (W3C; http://www.w3.org) to describe a specific set of technologies, including: the Extensible Markup Language (XML) to package and serialize data; SOAP (originally an acronym for ‘Simple Object Access Protocol’, but since version 1.2, just a capitalized name) and the HyperText Transfer Protocol (HTTP) to orchestrate communication between client and server; and the Web Service Description Language (WSDL) to describe the programmatic interface of the web service itself (http://www.w3.org/TR/wsdl). Today, the term web service has moved into common usage and its meaning has broadened considerably to encompass numerous other web-based mechanisms and approaches for providing remote programmatic service access. For providers and consumers alike, selecting an appropriate web service technology is a daunting task, requiring an in-depth understanding of numerous complex technological issues and implications. Although, on first inspection, some approaches to web service development appear to provide a relatively straightforward route via which providers may deploy their resources [e.g. REST (13) and traditional XML Remote Procedure Calls (XML-RPC; http://www.xmlrpc.com)], the consortium concluded that, on balance, and in the context of the life sciences, the strict guidelines, industry-supported validation tools, fault-tolerance and explicit description language associated with the original W3C definition provided benefits to the consumer that outweighed any short-term inconvenience to the Web service provider. Furthermore, it was considered that even the variety of options and configurations afforded by the W3C definition was too liberal to be practical; the decision was therefore made to adopt the more tightly specified subset of the W3C specification based on the profile defined by the Web Service Interoperability Organization (WS-I, a standards-defining consortium consisting of many of the major players in the IT industry including IBM, Microsoft, Oracle and Intel; http://www.ws-i.org). In addition, EMBRACE recommended the use of the emerging Semantic Annotations for WSDL (SAWSDL) (14) as a means of more richly describing the behaviour of services. More detailed reasoning behind these decisions is reported by Stockinger et al. (15) and ‘life-science friendly’ advice to the producers and consumers of web services is available in the project's Technology Recommendation documents, available online at the EMBRACE portal (http://www.embraceregistry.net/standards).
ENABLING SEMANTIC DESCRIPTION OF BIOINFORMATICS SERVICES
EMBRACE Data and Methods (EDAM) is an ontology for bioinformatics tools and data, consisting of a set of defined terms, relationships between these terms, and rules that govern the usage of the terms and their relationships. Terms for the initial version of the ontology were collected from analysis of the following tools and resources:
the EMBRACE web services;
SOAP-based services provided by the European Bioinformatics Institute (16);
the myGrid ontology (17); and
the NAR database and web server categorizations (2).
An initial version of the ontology is available in Open Biomedical Ontologies (OBO; http://www.obofoundry.org) format and uses the terms, relations and rules defined by the OBO Foundry (18). In its current form, EDAM consists of around 1750 terms, with associated definitions and 14 types of relations. The ontology and associated documentation is available at http://edamontology.sourceforge.net.
Terms in EDAM fall into the top-level categories shown in Table 2.
Table 2.
Examples of terms and their categories from the EDAM ontology
| Category | Example terms |
|---|---|
| Field | Sequence analysis; Alignment; Sequencing; Microarrays |
| Entity | Gene; Amino acid; Residue cluster; Active site; Atom–atom-interaction |
| Tool function | Sequence alignment; Pairwise sequence alignment, Sequence database search |
| Data resource | PDB database (19); GO ontology (20) |
| Data type | Sequence alignment; Sequence record; Comparison matrix; Phylogenetic tree |
| Identifier | PDB code; UniProtKB (21) accession number |
In the context of web services, EDAM plays two important roles. First, as a source of terms that can be added to WSDL files using the SAWSDL extension attributes, it enables the functions, inputs and outputs of a service to be semantically annotated, leading to improved searching from within repositories and better integration with workflow systems. Second, it enables the detailed data types exchanged by services to be more richly expressed, the benefits of which are described in the following section.
IMPROVING DATA EXCHANGE BETWEEN SERVICES
Although human-readable domain-specific text file formats have served a valuable purpose in bioinformatics for many years, the growing interest in interoperable web services has required more ‘computer friendly’ approaches to be developed. XML, which defines a format in which data of arbitrary complexity can be encoded, has become the de facto vehicle for inter-system communication (and indeed, forms the basis for the SOAP, WSDL and SAWSDL protocols mentioned previously). Although data records expressed in XML are typically more verbose and difficult for humans to read than their ‘flat file’ counterparts, numerous significant advantages accrue from its use. Of particular relevance here is the ability to use an XML Schema Document (XSD; http://www.w3.org/XML/Schema) to define the ‘grammar’ required during a particular exchange of data. The adherence of services to this grammar can automatically be validated by well proven industry standard software libraries, allowing tools to detect garbled or malformed inputs and outputs and preventing errors from propagating through to later stages in a service pipeline.
Detailed, fine-grained description of the exchange format by a dedicated XSD has multiple advantages both for the providers of web services and for their users. Data can be automatically validated without the need for bespoke software, increasing the security and making the service implementation less demanding. Conversion between different formats can be achieved reliably via the Extensible Stylesheet Language Transformation (XSLT; http://www.w3.org/TR/xslt) mechanism. Finally, the fine-grained components of the data types can be semantically annotated by terms from a controlled vocabulary such as EDAM.
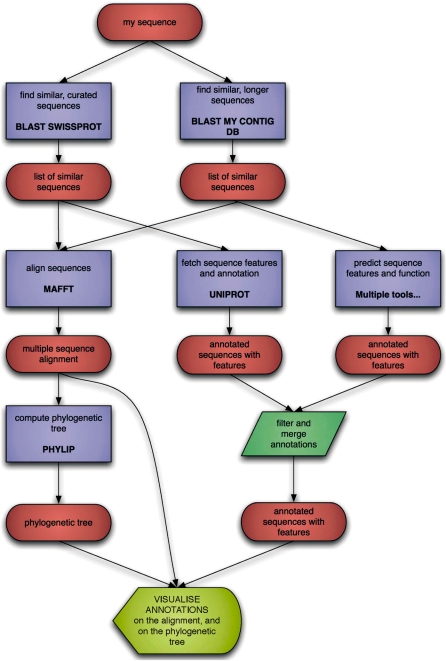
Maximum interoperability among the diverse bioinformatics web services located around the world can be achieved by using a common, canonical XML Schema. Defining the standard formats of the main data types, the canonical data model allows users to mix-and-match diverse services freely and without the need to write bespoke programs [sometimes referred to as ‘shims’ (22)] to transform to or from the myriad of legacy data formats. This makes the design of analytical workflows, scripts or programs much simpler, faster and cheaper, reducing the need for specialized personnel with advanced programming skills (Figure 1). On the side of the service providers, the common data model brings ready-made and semantically annotated data types that the developers of new services can, in many cases, use directly.
Figure 1.
An example of bioinformatics workflow, illustrating the flow of data (red ovals) through various services (blue rectangles). Without a common exchange format such as BioXSD, each edge in this graph would also require additional ‘converter' (or ‘shim’) processes to transform the data into the input formats required by the main services. This would more than double the technical complexity of the workflow for no additional scientific advantage.
In a similar vein to EDAM, BioXSD has been developed by analysing the existing web services, tools and various existing data formats, and by consulting the bioinformatics community. Initiated by the EMBRACE partners, BioXSD attempts to serve as the common data model for the most widely used, basic biological data exchanged with web services. The current version covers biological sequences, alignments and sequence annotations with both positional and non-positional features, and in addition, defines formats for references to databases or controlled vocabularies/ontologies, literature citations, bioinformatics accession numbers and identifiers. The core type definitions are accompanied by a number of generic helper types and recommended names for entities that are yet to be assigned ontological definitions. These everyday bioinformatics data types did not previously have any standard XML representations, despite representing both inputs and outputs of more than two-thirds of the web services developed by the project.
Transformers between the BioXSD and the main community textual or tabular formats are included in the BioXSD development, as well as the compatibility with the OpenBio libraries, such as BioPython and BioPerl.
The rules suggested in BioXSD align with a series of well-known resources, such as the Sequence Ontology, Gene Ontology or the BioSapiens Protein Feature Ontology. Examples of its use to describe GFF3 (http://www.sequenceontology.org/gff3.shtml) and UniProt Knowledgebase features is available at http://www.embraceregistry.net/BioXSD/.
SERVICE MONITORING
In order to give users an indication of service reliability, the project developed an ‘active registry’ capable of monitoring the behaviour of its web service collection, and of notifying consumers and service providers of any problems encountered (23). Providing basic classification and searching mechanisms, the registry has been available since October 2008, and has accumulated around 800 WSDL end point descriptions, representing considerably more than a 1000 bioinformatics ‘services’. As the EMBRACE project draws to a close, the data and functionality of the project's registry are being transferred to the BioCatalogue system (24) that provides more sophisticated curation, tagging, browsing and searching facilities and offers a sustainable long-term repository for the project's results.
CONCLUSION
EMBRACE has set the stage for bioinformatics web services. It did so by not only recommending standards and schemas for life science web services, but also by delivering in the region of a 1000 web services that are largely interoperable. Figure 2 shows the relative proportions of web service coverage, broken down into various high-level categories. This substantial collection will provide an incentive for future service providers to adopt the EMBRACE web service recommendations, because doing so makes their services interoperable with a rapidly increasing number of other services. We hope that this step forward in web service technology will allow bioinformaticians all over the world to keep up with the exponentially growing data volumes that the ‘omics revolution’ is producing; and we look forward to future NAR special volumes that hopefully will list many new databases, servers, services and facilities to facilitate research in the life sciences by making use of the web services found in the EMBRACE and BioCatalogue registries.
Figure 2.
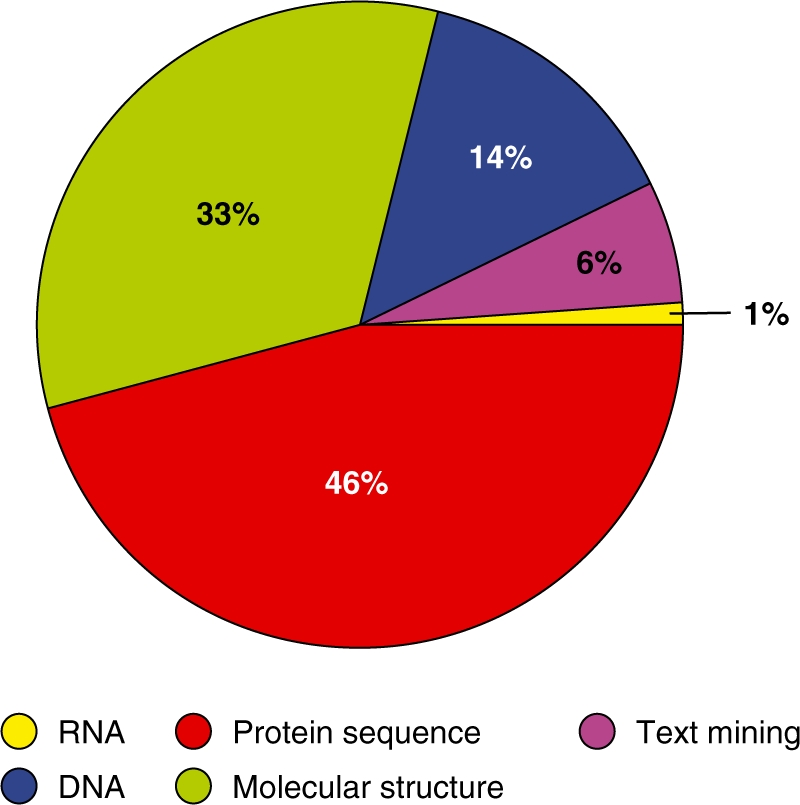
A pie chart showing the percentage of web services reported by the EMBRACE registry as belonging to various high-level categories.
FUNDING
This project reached a successful conclusion in no small part thanks to the persistent and patient input from its Project Manager, Kerstin Nyberg. Many scientists contributed by registering web services—their details are available via the project's registry; we especially acknowledge the input from the EU projects ENFIN and BioSapiens, and the numerous INB partners (http://www.inab.org) for registering very many services. Funding for open access charge: the publication charges will be paid from the EMBRACE budget via an account held at the EBI.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors gratefully acknowledge the contributions of Fred Marcus, Stephan Hohman, Rita Casadio, Matthew Woodwark, Jacques van Helden and Kay Hofmann for providing constant encouragement and stimulating discussions. European Commission within its FP6 Programme, under the thematic area ‘life sciences, genomics and biotechnology for health’ (contract number LHSG-CT-2004-512092 to EMBRACE project).
REFERENCES
- 1.Benson G. Nucleic Acids Research annual Web Server Issue in 2009. Nucleic Acids Res. 2009;37:W1–W2. doi: 10.1093/nar/gkv581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cochrane GR, Galperin MY. The 2010 Nucleic Acids Research Database Issue and online Database Collection: a community of data resources. Nucleic Acids Res. 2010;38:D1–D4. doi: 10.1093/nar/gkp1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Curcin V, Ghanem M, Guo Y. Web services in the life sciences. Drug Discov. Today. 2005;10:865–871. doi: 10.1016/S1359-6446(05)03481-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alonso G, Casati F, Kuno H, Machiraju V. Data-Centric Systems and Applications. Berlin/Heidelberg GmBH: Springer-Verlag; 2004. Web services: concepts, architectures and applications. [Google Scholar]
- 5.Wilkinson MD, Links M. BioMOBY: an open source biological web services proposal. Brief. Bionform. 2002;3:331–341. doi: 10.1093/bib/3.4.331. [DOI] [PubMed] [Google Scholar]
- 6.Djamal B, Dustar S, Seth A. Service Mashups: The new generation of web applications. IEEE Internet Comput. 2008;12:13–15. [Google Scholar]
- 7.Hull D, Wolstencroft K, Stevens R, Goble C, Pocock MR, Li P, Oinn T. Taverna: a tool for building and running workflows of services. Nucleic Acids Res. 2006;34:W729–W732. doi: 10.1093/nar/gkl320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Altintas I, Berkley C, Jaeger E, Jones M, Ludascher B, Mock S. Proceedings of 16th International Conference on Scientific and Statistical Database Management. 2004. Kepler: an extensible system for design and execution of scientific workflows; pp. 423–424. [Google Scholar]
- 9.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 10.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kopka J, Schauer N, Krueger S, Birkemeyer C, Usadel B, Bergmüller E, Dörmann P, Weckwerth W, Gibon Y, Stitt M, et al. GMD@CSB.DB: the Golm Metabolome Database. Bioinformatics. 2005;21:1635–1638. doi: 10.1093/bioinformatics/bti236. [DOI] [PubMed] [Google Scholar]
- 12.Vriend G. WHAT IF: a molecular modeling and drug design program. J. Mol. Graph. 1990;8:52–56. doi: 10.1016/0263-7855(90)80070-v. [DOI] [PubMed] [Google Scholar]
- 13.Richardson L, Ruby S. RESTful Web Services - Web services for the Real World. 2007. O'Reilly Media. [Google Scholar]
- 14.Kopecky J, Vitvar T, Bournez C, Farrell J. SAWSDL: Semantic Annotations for WSDL and XML Schema. IEEE Internet Comput. 2007;11:60–67. [Google Scholar]
- 15.Stockinger H, Attwood T, Chohan SN, Côté R, Cudré-Mauroux P, Falquet L, Fernandes P, Finn RD, Hupponen T, Korpelainen E, et al. Experience using Web services for biological sequence analysis. Brief. Bioinform. 2008;9:493–505. doi: 10.1093/bib/bbn029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pillai S, Silventoinen V, Kallio K, Senger M, Sobhany S, Tate J, Velankar S, Golovin A, Henrick K, Rice P, et al. SOAP-based services provided by the European Bioinformatics Institute. Nucleic Acids Res. 2005;33:W25–W28. doi: 10.1093/nar/gki491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wolstencroft K, Alper P, Hull D, Wroe C, Lord PW, Stevens RD, Goble CA. The myGrid ontology: bioinformatics service discovery. Int. J. Bioinform. Res. Appl. 2007;3:303–325. doi: 10.1504/IJBRA.2007.015005. [DOI] [PubMed] [Google Scholar]
- 18.Smith B, Ashburner M, Rosse C, Bard J, Bug W, Ceusters W, Goldberg LJ, Eilbeck K, Ireland A, Mungall CJ. The OBO Foundry: coordinated evolution of ontologies to support biomedical data integration. Nat. Biotechnol. 2007;25:1251–1255. doi: 10.1038/nbt1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kouranov A, Xie L, de la Cruz J, Chen L, WestBrook J, Bourne PE, Berman HM. The RCSB PDB information portal for structural genomics. Nucleic Acids Res. 2006;34:D302–D305. doi: 10.1093/nar/gkj120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ashburner M, Ball C, Blake A, Botstein JA, Butler D, Cherry H, Davis JM, Dolinski AP, Dwight K, Eppig SS, et al. Gene Ontology: tool for the unification of biology. Nat. Genetics. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Uniprot Consortium. The Universal Protein Resource (UniProt) in 2010. Nucleic Acids Res. 2010;38:D142–D148. doi: 10.1093/nar/gkp846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hull D, Stevens R, Lord P, Wroe C, Goble C. Proceedings of First Advanced Knowledge Technologies Workshop on Semantic Web Services (AKT-SWS04) KMi. Milton Keynes, UK: The Open University; 2004. Treating shimantic web syndrome with ontologies. [Google Scholar]
- 23.Pettifer S, Thorne D, McDermott P, Attwood T, Baran J, Bryne JC, Hupponen T, Mowbray D, Vriend G. An active registry for bioinformatics web services. Bioinformatics. 2009;25:2090–2091. doi: 10.1093/bioinformatics/btp329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bhagat J, Tanoh F, Nzuobontane E, Laurent T, Orlowski J, Roos M, Wolstencroft K, Stevens R, Pettifer S, Lopez R, et al. BioCatalogue : A universal catalogue of Web Services for the life sciences. Nucleic Acids Res. 2010 doi: 10.1093/nar/gkq394. [DOI] [PMC free article] [PubMed] [Google Scholar]