Abstract
The analysis of putative transcription factor binding sites in promoter regions of coregulated genes allows to infer the transcription factors that underlie observed changes in gene expression. While such analyses constitute a central component of the in-silico characterization of transcriptional regulatory networks, there is still a lack of simple-to-use web servers able to combine state-of-the-art prediction methods with phylogenetic analysis and appropriate multiple testing corrected statistics, which returns the results within a short time. Having these aims in mind we developed TransFind, which is freely available at http://transfind.sys-bio.net/.
INTRODUCTION
Searching for functional transcription factor binding sites in promoter regions has been a problem addressed for decades. Still, due to the short length of sequence motifs recognized by most vertebrate transcription factors and the excessively large non-coding DNA regions in these genomes, the annotation of binding sites in individual promoters is dominated by false predictions (1). A possible way to improve the specificity of the prediction for individual binding sites comes from considering the evolutionary conservation of a binding site between different species (2). The utility of such phylogenetic analysis has been demonstrated in finding distal enhancer regions (3). However, when concerned with proximal promoters, the advantage of using conservation is still debated. It has been shown that the conservation of individual binding sites differs strongly between different transcription factors (4). It could be argued that evolutionary pressure does not necessarily lead to conservation of an individual binding site, but rather to conservation of a general ability of the transcription factor to bind somewhere in the promoter and regulate the gene accordingly. Therefore, a possible way to alleviate this problem stems from approaches that predict the affinity of a transcription factor to entire promoter regions rather than to individual binding sites within them. Such affinity-based methods avoid the artificial separation between transcription factor binding sites and non-binding sites and were shown to emulate the in vivo binding behaviour more quantitatively than hit-based approaches (5–8). By ranking all promoters of a genome based on the predicted affinities, one can extract the likely candidate target genes of a particular transcription factor.
Statistical meta-analyses such as testing for enrichment of predicted sites or target genes in a set of coregulated genes, or in a set of genes with shared function, have been proven most useful to discover the transcription factors underlying the observed expression pattern (9,10). Such approaches draw upon the idea that a particular transcription factor likely regulates several genes at once. The corresponding predicted target genes should thus be enriched within the set of co-regulated genes.
To solve the problem of identifying the transcription factors regulating a given set of genes, a number of methods have been proposed that use either the annotation of discrete binding sites or continuous binding affinities (11–14). To determine the statistical significance of the results, most methods rely on computationally expensive resampling procedures or utilize discrete binding sites instead of affinity scores to predict the target genes of each individual transcription factor. In contrast, we present a method available through an easy-to-use web interface, which combines affinity measures (5) and support for phylogenetic analysis with rigorous statistics (9), and that returns the results within short time. Moreover, TransFind also features visualization of the GC- and CpG content of promoter sequences as well as the binding sites, which allows inspection and interpretation of the nucleotide composition of the input sequence sets and matrices. TransFind is freely accessible at http://transfind.sys-bio.net/.
TransFind SERVER
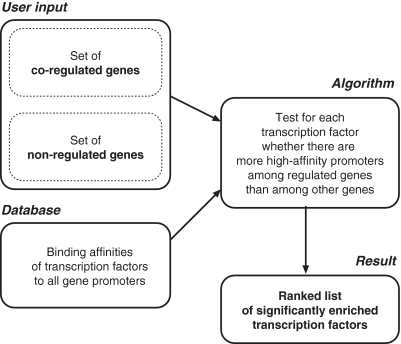
The TransFind service has been designed to conveniently solve a well-defined biological question: which transcription factor (TF) is a likely regulator of a given set of genes (in the following termed the positive set)? Such sets would for example consist of genes found to be up-regulated in a microarray experiment after a perturbation. In order to answer that question, we have set up an analysis pipeline and web server (illustrated in Figure 1).
Figure 1.
General overview of the algorithm.
When starting the analysis, the user is requested to provide a list of genes for the positive set. Lists are accepted in the form of any of the popular identifiers that are available as cross-references in Ensembl, such as Entrez Gene IDs or names, Ensembl identifiers or corresponding Affymetrix probe IDs. After submission, TransFind tests whether the list contains a significantly enriched number of putative target genes for any of the supported transcription factors. As targets, we define those genes of which the promoters display top-ranking affinities to the respective transcription factor.
The enrichment is measured with respect to another set of genes (the negative set) that by default contains all other genes of the organism. Often, it may be more appropriate to define only a subset of all genes to be the negative set. Such a list can be provided by the user. A typical user-defined negative set would consist of all genes that were found to be expressed in the microarray study, but do not show a change in expression between the conditions. Since genes can only be in either the positive set or in the negative set, TransFind automatically excludes genes present in the positive set from the negative set.
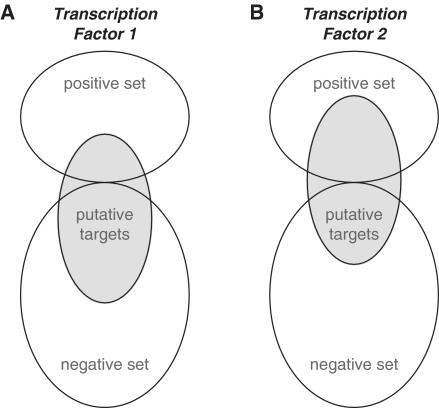
We use Fisher's exact test (Figure 2) to quantify enrichment of putative high-affinity targets of a transcription factor in the positive set. Since we test for the enrichment of a multitude of transcription factor matrices, correcting for multiple testing is an issue. We have previously established an analytical approach (9) to determine the false discovery rate (FDR).
Figure 2.
TransFind identifies transcription factors with significantly enriched numbers of predicted targets in the regulated gene set with respect to an unregulated set. (A) The putative targets of factor 1 distribute randomly among the regulated genes (positive set) and non-regulated genes (negative set). (B) In contrast, the top targets of factor 2 are strongly enriched in the regulated genes.
Once corrected for multiple testing, TransFind reports the results as a table of transcription factor matrices ranked by the enrichment of their predicted targets in the positive set. By default, only the significant results are shown. We define any transcription factor matrix as significant if the corresponding FDR is <0.05, which limits the fraction of false predictions to 5%. Additionally, a link is provided to a table listing the detailed results for all analysed transcription factor matrices.
An example of the output which includes the supporting statistical details are is given in Table 1. For each transcription factor matrix, the server reports how many of the genes from the positive set have a promoter with strong predicted affinity to the factor. This can be compared to the number of predicted promoters with high affinity to the factor in the negative set. The results of the statistical test are provided in further columns, including the corresponding P-value of the Fisher's exact test, FDR and expected numbers of false positives (FPs). TransFind also displays the logo of the sequence motif that is recognized by the transcription factor (15). It is possible to obtain the results in a simple text format or in the XML format containing additional details on the mapping of the input gene identifiers.
Table 1.
An example of a TransFind result showing all significant transcription factor matrices predicted to regulate a known set of 51 c-myc targets
| Rank | TF matrix | P-value | FDR | FP | Hits in positive set ES (%) | Hits in negative set ES (%) |
|---|---|---|---|---|---|---|
| 1 | V$NMYC_01 / N-Myc | 0.000001 | 0.000001 | 0.000001 | 11 (22.45) | 489 (0.98) |
The top 500 predicted targets of each transcription factor were analysed. The set of matrices was limited to the most informative per Transfac factor. Following the matrix and factor names, the P-value, FDR and expected number of FP are reported. Furthermore, the number of high-affinity matches for the factor in the positive set and in the negative set are shown, together with their relative abundance.
To facilitate rapid calculation, we utilize arrays containing precalculated affinity scores to all promoter regions. Additionally, we provide precalculated scores for phylogenetically conserved regions. These scores were calculated for all genes that have orthologues in another selected vertebrate species, by taking either the average or the minimal affinity from both organisms. This approach allows detecting promoters where the individual binding sites might not be conserved, but for which the transcription factor still binds with high affinity in both organisms.
The strongest enrichment for functional transcription factor binding sites has been previously found to lie within the first few 100 base pairs upstream of the transcription start site (16). Therefore, our default promoter set contains promoter sequences ranging from 300 nt upstream of the gene start to 100 nt downstream of the gene start annotated in Ensembl. In addition, users may select another promoter set that consists of sequences spanning from 800 nt upstream to 200 nt downstream of the gene start.
We provide three different sets of transcription factor matrices. First, the user may select a complete set of vertebrate transcription factor matrices that are contained in the Transfac database (17). As this complete set of matrices is highly redundant, the results are often difficult to interpret (18). Therefore, we created a reduced set of matrices containing only a single, the most informative matrix for each vertebrate Transfac transcription factor. The third set of matrices has been downloaded from the rapidly developing and free transcription factor binding profile database Jaspar (19).
PERFORMANCE EVALUATION
Based on published experimental data, we collected six sets of target genes for the following transcription factors: c-myc (20), E2F (21), NFκB (22), Hif1a (23), Hnf4 (http://www.sladeklab.ucr.edu/hnf43.pdf) and Ets1 (24). Table 1 illustrates the output from TransFind obtained for the set of c-myc-regulated genes. The enrichment has been computed for the overlap between the 51 input genes and the top 500 targets for each TF as predicted from the affinities for short promoters of length 400 bp. For this data set, TransFind predicts the myc-related transcription factor N-Myc as strongly associated.
Also, for each of the literature test sets of Hif1a, Hnf4 and E2F targets. TransFind correctly identifies the corresponding TF matrices as most significantly enriched. In case of E2F and the complete set of Transfac matrices, one obtains all 21 variants of the E2F matrix as significantly associated. In contrast, for the reduced set of matrices, the algorithm returns only three of the most informative matrices of the E2F transcription factors. The set of E2F targets also illustrates that TransFind warns the user when an unusual CpG composition is found in the promoters of the provided genes. The histograms displaying bias of the CpG- or GC composition might be then inspected (see below). Similarly, for the NFκB target set and the reduced set of matrices, TransFind reports four similar NFκB motifs as well as the related factors c-REL and HMG as likely regulators of the set. In comparison, TransFind returns 21 redundant NFκB matrices when utilizing the full matrix set.
Finally, TransFind reports no significantly associated TF for the set of Ets1 targets when using the standard FDR cut-off of 0.05, indicating that none of the transcription factor matrices has enriched affinity for the promoters of the targets of Ets1. However, when the significance cut-off is relaxed, the correct TF motif is recovered at position six after several matrices corresponding to the transcription factor AP1 as well as a matrix from BACH1.
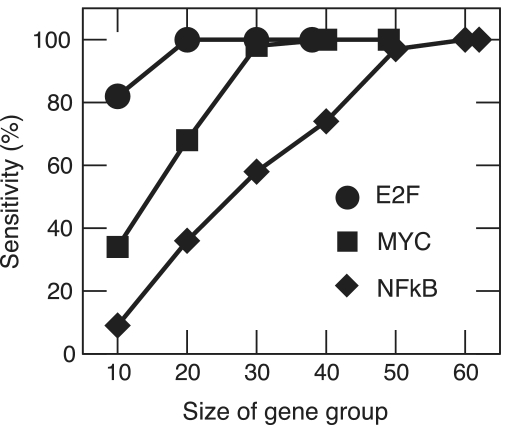
We verified how many genes are sufficient to identify an associated transcription factor. Hence, we generated random subsets of the literature-derived E2F, NFκB and c-myc gene sets of different sizes and ran TransFind on them. We then defined sensitivity as the fraction of runs for which the correct transcription factor was recovered. The results are shown in Figure 3. It turns out that minimum positive set size depends on the transcription factor, however, for the set of E2F targets even small subsets of 10 genes are often sufficient.
Figure 3.
Sensitivity of TransFind for different sizes of the positive gene set. We used TransFind on random subgroups of literature-derived E2F, c-myc and NFκB target genes in order to determine minimum number of genes sufficient for correct identification of the regulating transcription factor. We defined sensitivity as the fraction of randomly selected subgroups, which resulted in a significant prediction of the respective transcription factor. We used default TransFind settings (500 top affinities, subset of Transfac matrices with highest information content).
In summary, TransFind provides predictions in agreement with biological knowledge in five out of six available experimental data sets. These predictions are robust with respect to changes of the parameters. The analyses can be easily repeated with the help of example buttons loading the experimental data sets.
In order to estimate TransFind performance, we prepared gene sets annotated with the same Gene Ontology term. By limiting the sets to genes annotated with terms close to the root of the ontology (not more than 2 steps away from biological process, molecular function or cellular location) and taking only sets with more than 10 genes, we constructed 397 gene sets that we systematically analyse. We expect that many of these sets are not correlated in their expression. However, some of these sets will be coregulated and therefore consist of genes that share transcription factors that regulate them. Thus, these sets can be used to systematically analyse the performance of TransFind with different parameter settings, but it has to be kept in mind that the sensitivity will be under-estimated. We found that (e.g. using the top 200 genes as targets and the default promoter set of length 400 nt) 23% of the gene sets were predicted to be regulated by at least one transcription factor. One hundred and twenty-one out of the non-redundant set of 217 transcription factor matrices were predicted to regulate at least one set, suggesting that it is not a small group of matrices that dominates the analysis.
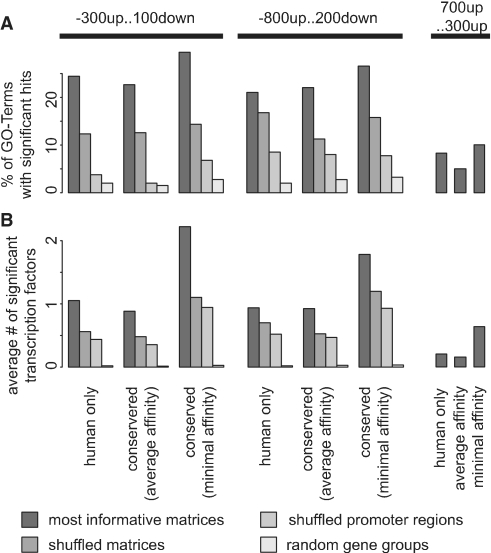
Next, we investigated whether phylogenetic information improves the prediction performance. When using average affinity for human and mouse, we observe a mild reduction in the number of predictions. In contrast, when using minimal affinity, more regulating transcription factors are predicted. We summarize these results in Figure 4.
Figure 4.
Performance of TransFind. We defined 397 sets of genes which were annotated with top Gene Ontology terms and searched for enriched putative transcription factor targets using different parameters. Results for original data (dark grey) are compared to results for shuffled matrices or promoter sequences (medium grey) or to random gene sets of the same size distribution (light grey). Panel A shows the fraction of GO sets with at least one significant factor and panel B shows the average number of discovered factors in a GO set.
The fraction of false predictions was estimated using different randomization scenarios. First, we shuffled positions within each transcription factor matrix. This method gave the highest estimations of the fraction of falsely predicted factors. However, we interpret this result as an overestimation, since a shuffled matrix is often by chance very similar to the original matrix (especially for repetitive or skewed matrices). Next, we ran TransFind on promoter sets with nucleotide sequences shuffled within each promoter. The resulting fraction of false predictions was <5%. If conservation was taken into account by using average affinities between human and mouse orthologues, the fraction of false predictions was even smaller. In contrast, when using minimal affinities, the number of false predictions increased as did the number of predictions. Finally, we also sampled random gene sets with the same size distribution as for the reference 397 gene sets. For only about 2% of these random gene sets was any significant factor found, independent of whether conservation was used or not. Overall, the results suggest that for about 5–8% of submitted gene set sets, false predictions are returned. Choosing minimal affinity, the results show more predictions but at the cost of increased number of false predictions. In contrast, by using average affinity the amount of false predictions can be reduced.
Based on the results for shuffled promoters, one could deduce that false predictions occur mainly because of biased nucleotide composition of the promoter sequences. In the analysis of the 397 Gene Ontology groups, we identified 17 groups (5%) with promoters displaying either unusually high or low CpG- or GC content. Therefore, TransFind additionally provides a test for gene sets with biased promoters and displays a warning when such a significant enrichment occurs. The user should then carefully consider whether the transcription factors are found only due to the overall promoter composition, and if other modes of regulation such as epigenetic regulation should be investigated.
To assess whether the short core promoter regions are sufficient to predict the regulating factors, we repeated the analysis on a set of promoters of length 1000 nt, which showed no better performance. This observation is also supported by the comparison of the short core promoter regions directly upstream of them (from 700 nt upstream to 300 nt upstream of the gene start sites). Predictions for these shifted regions were hardly better than the predictions for the shuffled promoter sequences.
To conclude, our systematic analyses suggest that taking the short core promoters (300 nt upstream to 100 nt downstream of gene start) together with minimum affinity in mouse and human provides the best sensitivity without many false predictions.
IMPLEMENTATION DETAILS
Extraction of promoter sequences
The Ensembl database (25) in its current version 57 is used as the source of genomic sequences and corresponding gene annotations. We extracted two sets of putative promoters: short—covering the range of 300 nt upstream to 100 nt downstream and long—from 800 nt upstream to 200 nt downstream of the most upstream transcription start site.
Prediction of binding affinities
We use a previously published approach (5) to calculate binding affinities of a transcription factor to a gene promoter. Since this calculation is time-consuming, we precalculated arrays of the affinities for all promoters and all transcription factors. We used either all vertebrate transcription factor matrices available in Transfac (17) version 2009.4 or the non-redundant vertebrate matrices from Jaspar core (19). Additionally, since Transfac is highly redundant, we used a subset of matrices, where for each transcription factor we select the matrix with the highest information content. Apart from the calculation of the different transcription factor binding affinities, we determine the GC- and CpG content (26) of each promoter sequence and subsequently use these values along with the affinities in the statistical test.
Phylogenetic analysis
TransFind provides an option to consider only phylogenetically conserved regulation. In this mode, affinity of a factor to a gene is combined with affinity of the same factor to the gene orthologue in a selected organism. Either minimum or average of the two affinities is computed. In the minimum mode, genes with high affinities in both organisms are top ranked.
Identifier mapping
In the first step of analysis, TransFind maps provided input gene names to the genes present in the affinity arrays. The mapping tables are constructed based on the cross-reference annotations available for each gene in Ensembl. If an input identifier is mapped to several genes, all of them are included in further analysis. Multiple occurrences of the same gene are unified automatically. To guarantee that there is no overlap between positive and negative sets, all genes common to both sets are excluded from the negative set of genes. Thus, users can simply paste their gene sets without any prior conversion directly into to the input form and immediately start TransFind. If no negative set is provided, all genes of the genome that are not in the positive set are taken.
Statistical analysis
For each transcription factor we identify a chosen number of top-ranking genes based on precalculated affinities. Subsequently, using a multiple testing corrected Fisher's exact test we check whether there exists a transcription factor with top-ranked genes enriched among the submitted positive set of genes when compared to the negative set of genes (27).
Web server
The results are presented in a form of a ranked table, with links to pages providing more information about positional frequency matrices and corresponding transcription factors. We also display sequence logos to visualize the positional frequency matrices and provide histograms of the GC- and CpG content of the positive and negative sequence sets. Internally, all computed results are kept in XML format. The functionality of the web server is embedded into a content management system (Joomla), which allows to efficiently manage multiple sessions and to rapidly change the description and help pages to update the web site and incorporate user suggestions.
CONCLUSIONS
We have implemented an easy-to-use web server that allows to predict transcription factors regulating a set of genes using state-of-the-art methods. The method has been successfully evaluated on various data sets. The web site is free and open to all users at http://transfind.sys-bio.net/ and there is no login requirement.
FUNDING
German National Genome Research Network (NGFN-Plus, grant 01GS0815); German Federal Ministry of Education and Research (BMBF) (grant FORSYS-Partner); Deutsche Forschungsgemeinschaft (grant SFB 618); European Commission (grant number CancerSys HEALTH-F4-2008-223188); International Research Training Group (IRTG) for Genomics and Systems Biology of Molecular Networks. Funding for open access charge: NGFN-Plus (grant 01GS0815).
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We would like to thank Ralf Mrowka for valuable comments and suggestions.
REFERENCES
- 1.Wasserman WW, Sandelin A. Applied bioinformatics for the identification of regulatory elements. Nat. Rev. Genet. 2004;5:276–287. doi: 10.1038/nrg1315. [DOI] [PubMed] [Google Scholar]
- 2.Dieterich C, Rahmann S, Vingron M. Functional inference from non-random distributions of conserved predicted transcription factor binding sites. Bioinformatics. 2004;20(Suppl. 1):i109–i115. doi: 10.1093/bioinformatics/bth908. [DOI] [PubMed] [Google Scholar]
- 3.Mrowka R, Steege A, Kaps C, Herzel H, Thiele BJ, Persson PB, Blüthgen N. Dissecting the action of an evolutionary conserved non-coding region on renin promoter activity. Nucleic Acids Res. 2007;35:5120–5129. doi: 10.1093/nar/gkm535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sauer T, Shelest E, Wingender E. Evaluating phylogenetic footprinting for human-rodent comparisons. Bioinformatics. 2006;22:430–437. doi: 10.1093/bioinformatics/bti819. [DOI] [PubMed] [Google Scholar]
- 5.Roider HG, Kanhere A, Manke T, Vingron M. Predicting transcription factor affinities to DNA from a biophysical model. Bioinformatics. 2007;23:134–141. doi: 10.1093/bioinformatics/btl565. [DOI] [PubMed] [Google Scholar]
- 6.Ward LD, Bussemaker HJ. Predicting functional transcription factor binding through alignment-free and affinity-based analysis of orthologous promoter sequences. Bioinfonnatics. 2008;24:i165–i171. doi: 10.1093/bioinformatics/btn154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tanay A. Extensive low-affinity transcriptional interactions in the yeast genome. Genome Res. 2006;16:962–972. doi: 10.1101/gr.5113606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Granek JA, Clarke ND. Explicit equilibrium modeling of transcription-factor binding and gene regulation. Genome Biol. 2005;6:R87. doi: 10.1186/gb-2005-6-10-r87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Blüthgen N, Kielbasa SM, Herzel H. Inferring combinatorial regulation of transcription in silico. Nucleic Acids Res. 2005;33:274–279. doi: 10.1093/nar/gki167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tullai J, Schaffer M, Mullenbrock S, Kasif S, Cooper G. Identification of transcription factor binding sites upstream of human genes regulated by the phosphatidylinositol 3-kinase and mek/erk signaling pathways. J. Biol. Chem. 2004;279:20167–20177. doi: 10.1074/jbc.M309260200. [DOI] [PubMed] [Google Scholar]
- 11.Sui SJH, Fulton DL, Arenillas DJ, Kwon AT, Wasserman WW. OPOSSUM: integrated tools for analysi of regulatory motif over-representation. Nucleic Acids Res. 2007;35:W245–W252. doi: 10.1093/nar/gkm427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Roider HG, Manke T, O'Keeffe S, Vingron M, Haas SA. Pastaa: identifying transcription factors associated with sets of co-regulated genes. Bioinformatics. 2009;25:435–442. doi: 10.1093/bioinformatics/btn627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chang L-W, Fontaine BR, Storno GD, Nagarajan R. Pap: a comprehensive workbench for mammalian transcriptional regulatory sequence analysis. Nucleic Acids Res. 2007;35:W238–W244. doi: 10.1093/nar/gkm308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Frith MC, Fu Y, Yu L, Chen J-F, Hansen U, Weng Z. Detection of functional DNA motifs via statistical over-representation. Nucleic Acids Res. 2004;32:1372–1381. doi: 10.1093/nar/gkh299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schneider TD, Stephens RM. Sequence logos: a new way to display consensus sequences. Nucleic Acids Res. 1990;18:6097–6100. doi: 10.1093/nar/18.20.6097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Roider HG, Lenhard B, Kanhere A, Haas SA, Vingron M. CpG-depleted promoters harbor tissue-specific transcription factor binding signals–implications for motif overrepresentation analyses. Nucleic Acids Res. 2009;37:6305–6315. doi: 10.1093/nar/gkp682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Matys V, Kel-Margoulis O, Fricke E, Liebich I, Land S, Barre-Dirrie A, Reuter I, Chekmenev D, Krull M, Hornischer K, et al. Transfac and its module transcompel: transcriptional gene regulation in eukaryotes. Nucleic Acids Res. 2006;34:D108–D110. doi: 10.1093/nar/gkj143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kielbasa SM, Gonze D, Herzel H. Measuring similarities between transcription factor binding sites. BMC Bioinformatics. 2005;6:237. doi: 10.1186/1471-2105-6-237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Portales-Casamar E, Thongjuea S, Kwon AT, Arenillas D, Zhao X, Valen E, Yusuf D, Lcnhald B, Wasserman WW, Sandelin A. Jaspar 2010: the greatly expanded open-access database of transcription factor binding profiles. Nucleic Acids Res. 2010;38:D105–D110. doi: 10.1093/nar/gkp950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fernandez PC, Frank SR, Wang L, Schroeder M, Lill S, Greene J, Cocito A, Amati B. Genomic targets of the human c-myc protein. Genes Dev. 2003;17:1115–1129. doi: 10.1101/gad.1067003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bracken AP, Ciro M, Cocito A, Helin K. E2F target genes: unraveling the biology. Trends Biochem. Sci. 2004;29:409–417. doi: 10.1016/j.tibs.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 22.Wu JT, Krai JG. The NF-κb/IκB signaling system: a molecular target in breast cancef therapy. J. Surg. Res. 2005;123:158–169. doi: 10.1016/j.jss.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 23.Semenza GL. Targeting HIF-I for caricer therapy. Nat. Rev. Cancer. 2003;3:721–732. doi: 10.1038/nrc1187. [DOI] [PubMed] [Google Scholar]
- 24.Sementchenko YI, Watson DK. Ets target genes: past, present and future. Oncogene. 2000;19:6533–6548. doi: 10.1038/sj.onc.1204034. [DOI] [PubMed] [Google Scholar]
- 25.Hubbard TJP, Aken BL, Ayling S, Ballester B, Beal K, Bragin E, Brent S, Chen Y, Clapham P, Clarke L, et al. Ensembl 2009. Nucleic Acids Res. 2009;37:D690–697. doi: 10.1093/nar/gkn828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Saxonov S, Berg P, Brutlag DL. A genome-wide analysis of cpg dinucleotides in the human genome distinguishes two distinct clases of promoters. Proc. Natl. Acad. Sci. USA. 2006;103:1412–1417. doi: 10.1073/pnas.0510310103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Blüthgen N, Brand K, Cajavec B, Swat M, Herzel H, Beule D. Biological profiling of gene groups utilizing gene ontology. Genome Inform. 2005;16:106–115. [PubMed] [Google Scholar]