Abstract
A web service for analysis of protein structures that are sequentially or non-sequentially similar was generated. Recently, the non-sequential structure alignment algorithm GANGSTA+ was introduced. GANGSTA+ can detect non-sequential structural analogs for proteins stated to possess novel folds. Since GANGSTA+ ignores the polypeptide chain connectivity of secondary structure elements (i.e. α-helices and β-strands), it is able to detect structural similarities also between proteins whose sequences were reshuffled during evolution. GANGSTA+ was applied in an all-against-all comparison on the ASTRAL40 database (SCOP version 1.75), which consists of >10 000 protein domains yielding about 55 × 106 possible protein structure alignments. Here, we provide the resulting protein structure alignments as a public web-based service, named GANGSTA+ Internet Services (GIS). We also allow to browse the ASTRAL40 database of protein structures with GANGSTA+ relative to an externally given protein structure using different constraints to select specific results. GIS allows us to analyze protein structure families according to the SCOP classification scheme. Additionally, users can upload their own protein structures for pairwise protein structure comparison, alignment against all protein structures of the ASTRAL40 database (SCOP version 1.75) or symmetry analysis. GIS is publicly available at http://agknapp.chemie.fu-berlin.de/gplus.
INTRODUCTION
The comparison of native three-dimensional (3D) protein structures is one of the most essential strategies of structural biology. It is the structure, which determines a protein’s biochemical functions, whereby structural similarity of one protein to another is an indication for similar function and/or evolutionary relation. Although the protein structure is fully determined by the protein’s amino acid sequence, protein structure analysis is often superior to sequence-based approaches. This is particular true, if the sequence identity of the analyzed protein pair is low. It relates to the fact that protein structures are evolutionary more conserved than protein sequences and consequently the universe of protein structures is less complex than the space of protein sequences. Therefore, protein pairs with distantly related sequences might still share a common protein fold, since protein structures are generally more conserved than protein sequences. A protein sequence of only ten amino acids e.g. already yields a set of 2010 possible sequences, which highly exceeds the number of protein folds observed in nature.
Several algorithms have been developed in recent years to solve the problem of protein structure comparison in various approximations e.g. DaliLite (1), K2 (2), CE (3) and TM-align (4). Although these methods are very successfully, most of them are restricted or biased considering preferentially the similarity of protein structures possessing the same connectivity of secondary structure elements (SSEs; i.e. α-helices and β-strands) as defined by the polypeptide chain. Only a few methods are available that allow for non-sequential protein structure alignments e.g. MASS (5), TOPOFIT (6), SCALI (7), GANGSTA (8,9) and others (10,11). Recently, GANGSTA+ (12) was introduced and we presented several applications such as the detection of non-sequential structural analogs for novel protein folds (12), the detection of symmetric (13) and circular permuted (14,15) protein structures in the protein databases and a large-scale evaluation of all-against-all sequence and structure alignments (16).
Here, a database of protein structure alignments (GIS) was generated by applying GANGSTA+ in an all-against-all comparison with more than 10 000 protein structures of the ASTRAL40 (SCOP version 1.75) database. A user-friendly web service has been made publicly available and enables users to select, visualize and analyze the generated protein structure alignments with regard to structure and sequence conservation. In addition to the presented web services, we hereby provide the GANGSTA+ source code as free download (General Public License). The website is free and open to all users and there is no login requirement.
MATERIALS AND METHODS
The non-sequential protein structure alignment algorithm GANGSTA+ was used in the present study. GANGSTA+ aligns protein structures hierarchically starting with an alignment of SSE (first stage), where only α-helices and β-strands are considered as SSEs. Non-sequential structure alignment is facilitated, since loops and coils connecting the SSEs are ignored. GANGSTA+ uses a combinatorial approach to optimize the SSE assignment. For the highest ranked SSE assignments preliminary alignments on the residue level are performed (second stage) using energy minimization with attractive soft interactions between Cα atom pairs belonging to different proteins. In stage three, this preliminary structure overlay is used to assign the Cα atoms of both proteins on a 3D grid and using the closeness of Cα atom pairs found from the grid for a new more accurate and complete SSE assignment. Finally the assignment on residue level is repeated aligning now also residues, which belong to loops and coils if possible. For a more detailed algorithmic description of the optimization strategies employed in GANGSTA+, see the supplemental material of (12).
Content of the structure alignment database
We applied GANGSTA+ in an all-against-all protein structure comparison with the ASTRAL40 (SCOP version 1.75) database, which contains 10 444 out of 10 511 structures of protein domains with more than two SSEs to generate the protein structure alignment database available as web service. Hence, about 55 × 106 possible protein structure pairs were analyzed for structural similarity. GANGSTA+ is capable to detect sequential and non-sequential structural similarities between protein pairs and constraints can be set to obtain exclusively alignments with sequential or non-sequential SSE order. Here, the alignments were performed without constraints on SSEs order yielding alignment results in sequential or non-sequential SSE order. However, SSE pairs were not aligned in reverse orientation to each other (C-terminus of one SSE on the N-terminus of the other). Protein structures with reversely oriented SSEs were considered recently to discriminate evolutionary related circular permuted protein structure pairs from those, which occurred by chance (14). For each protein pair, only the structure alignment with the largest number of aligned residues and root mean square deviation (RMSD) below ∼4 Å Cα was kept.
Each protein structure alignment result is stored together with a set of six descriptors. These descriptors are: (i) ‘number of aligned SSEs’; (ii) ‘number of aligned residues’; (iii) fraction of aligned residues (equivalence) relative to the smaller of both proteins; (iv) Cα RMSD (RMSD) of aligned residues; (v) for protein structure alignment with all SSE pairs assigned sequentially (sequential) and (vi) circular permuted protein structures (circular permuted) (i.e. protein structure alignments with exactly one break in the sequential order of assigned SSE pairs without considering gaps). A detailed analysis regarding the detection of circular permuted protein structures with GANGSTA+ has been reported previously (14).
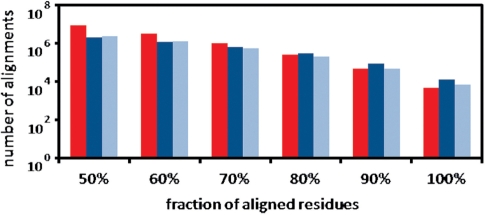
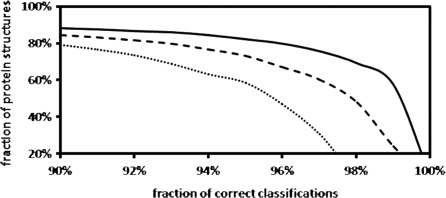
In about 13 × 106 protein structure alignments, GANGSTA+ succeeded to align at least 50% of the residues of the smaller of the two considered protein structures with a Cα RMSD below ∼4 Å. This number includes 2 012 002 ‘sequential’ and 2 400 789 ‘circular permuted’ protein structure alignments, but the vast majority of aligned protein pairs are non-sequential. Figure 1 shows the distribution of the fraction of aligned residues as detected by GANGSTA+. The fraction of aligned residues (equivalence) is determined with respect to the smaller protein of each aligned protein structure pair. Table 1 illustrates the classification performance of GANGSTA+ with respect to the SCOP classification scheme. The performance is shown according to the number of residues (≥60, ≥80 and ≥100) contained in the classified protein structures. For each protein structure, only the alignment with the largest number of aligned residues below ∼4 Å Cα RMSD and ‘equivalence’ (fraction of aligned residues) larger than 80% were considered. Figure 2 shows the detailed results of the fold recognition ranked by ‘equivalence’ according to the SCOP classification scheme.
Figure 1.
Number of protein structure alignments detected with GANGSTA+ in an all-against-all comparison of the ASTRAL40 database (SCOP version 1.75), consisting of 10 444 protein structures with more than two SSEs. The fraction of aligned residues (Cα atom RMSD below ∼4 Å) is determined with regard to the number of residues in the smaller protein of the aligned protein structure pair. The red bar corresponds to non-sequential structure alignments. The dark blue bar represents the ‘sequential’ and the light blue bar the ‘circular permuted’ structure alignments only.
Table 1.
Protein classification based on structural similarity according to the SCOP classification scheme
| Residuesa | Familyb (%) | Superfamilyb (%) | Foldb (%) | Totalc |
|---|---|---|---|---|
| ≥100 | 85.5 | 96.1 | 97.7 | 5545 |
| ≥80 | 81.7 | 93.0 | 95.2 | 6564 |
| ≥60 | 78.0 | 89.8 | 92.7 | 7333 |
Results obtained with GANGSTA+ applied to the ASTRAL40 (SCOP version 1.75) dataset of 10 444 proteins with more than two SSEs. Only the highest ranked alignments per protein structure with ‘equivalence’ larger than 80% were considered, resulting in an average coverage of 74%.
aMinimum total number of residues in the classified protein structures.
bFraction of classification results that agree with the SCOP classification scheme.
cTotal number of classified protein structures.
Figure 2.
Fold class recognition according to the SCOP classification scheme. Results obtained with GANGSTA+ applied to the ASTRAL40 (SCOP version 1.75) dataset of 10 444 proteins with more than two SSEs. For each protein structure, the alignment with the largest number of aligned residues was considered. The ‘equivalence’ was used to rank the resulting protein structure alignments. Protein structures with at least 60, 80 and 100 residues are shown as dotted, dashed and solid lines, respectively.
APPLICATIONS
In the following section, we illustrate several applications provided by the presented web service. First, users are able to browse and visualize the 3D structure alignment results at http://agknapp.chemie.fu-berlin.de/gplus. Additionally, our web page enables users to upload and align own protein structures against the ASTRAL40 database, to do pairwise comparisons or to analyze the intrinsic molecular symmetry of protein structures by non-trivial self-alignment. A detailed analysis of protein structures containing intrinsic rotational symmetries has been reported previously (13). We provide an online query script, which enables the integration of the GIS protein structure alignments in external software applications or web services. An example for such an application is given by STRAP (available at http://www.charite.de/bioinf/strap/), which is a JAVA-based graphical user interface for structure-based analysis of multiple protein sequence alignments (17).
Pairwise comparison of protein structures
Given a pair of protein structures in PDB file format (18), users can apply GANGSTA+ directly through our web service. Initially, users may provide their email address (to receive a query notification) and two protein structure files from the user’s local drive for upload. Afterwards, the resulting structure alignment can be viewed online or alternatively be downloaded as PDB file for local inspection.
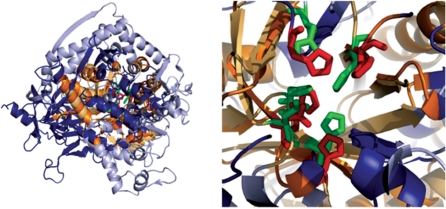
Here, we demonstrate the application of GANGSTA+ through our web service by aligning the protein structures of adenosine deaminase (PDB entry 2A3L, chain A) (19) and urease (PDB entry 1IE7, chain C) (20), which were recently in the focus of a review article by Hasegawa and Holm (21). These two proteins possess similar active sites, consisting of five conserved residues, responsible for metal-binding and catalytic activities. These residues are His137, His139, His249, His275, Asp363 in urease and His391, His393, His659, His681, Asp736 in adenosine deaminase. The authors applied 32 different structure alignment methods and analyzed the obtained structure alignment results with regard to the correct detection of the active site, respectively, the correct superposition of each of the five conserved residue pairs. Only six out of the 32 considered structure alignment methods [SSAP (22), LGA/GDT (23), TOPOFIT (6), GASH (24), PPM (25) and DaliLite (1)] were able to correctly pairwise superimpose all five conserved residues in the resulting protein structure alignment. Figure 3 shows the structure alignment result for adenosine deaminase (PDB id 2A3L, chain A) and urease (PDB id 1IE7, chain C) obtained by applying GANGSTA+ through our web service [visualized with PyMOL (26)]. Similar to the six successful algorithms mentioned above, GANGSTA+ generated the correct superposition of all five functionally relevant residue pairs.
Figure 3.
Left: protein structure alignment of adenosine deaminase (PDB code 2A3L, chain A, not aligned parts in light blue) (19) and urease (PDB code 1IE7, chain C, not aligned parts in dark blue) (20) generated with GANGSTA+ (12) at http://agknapp.chemie.fu-berlin.de/gplus and visualized with PyMOL (26). In the alignment mode using the PDB files, explicitly GANGSTA+ aligned 152 residues at 3.3 Å Cα RMSD shown in light, respectively, dark orange for adenosine deaminase and urease, respectively. These two proteins belong to different superfamilies according to SCOP (27), but possess a common metal binding motif. The five conserved residues pairs, responsible for metal-binding and catalytic activity are highlighted in green for adenosine deaminase and red for urease. The conserved residues are His391, His393, His659, His681 and Asp736 in adenosine deaminase and His137, His139, His249, His275 and Asp363 in urease. The structure alignment result generated with GANGSTA+ correctly superimposes the five conserved residues. Right: same as left, but with an enlarged view on the active site.
For pairwise protein structure alignments, one has two options: (i) providing just the PDB ids of the proteins the structures are taken from the locally available ASTRAL40 database of protein domains and (ii) alternatively one can provide the PDB structure file explicitly. In the former case, the structure database may not contain the structures of the proteins for which an alignment was requested. Then, the protein domain with the most similar sequence is taken from the database instead. In each case, the SCOP ids of the protein domains, which were actually used for the alignment, are given and warnings are issued, if the protein domains differ from the originally given proteins. Using explicitly, the two protein structures files from the PDB (2A3L, chain A and 1IE7, chain C) 152 residues were aligned at 3.3 Å Cα RMSD. If alternatively one provides the PDB ids of the two proteins, only an alignment with 141 residues at 3.2 Å Cα RMSD is obtained.
Using the protein structure alignment browser
Given a protein structure in PDB file format (18), users can apply GANGSTA+ to search for similar entries in the ASTRAL40 (SCOP version 1.75) database consisting of more than 10 000 protein structures. Initially, users may provide their email address (to receive a query notification) and one protein structure file from the user’s local drive for upload. Depending on the number of SSEs in the target protein structure, the calculation takes between 20 and 100 min on the currently used single-core AMD OPTERON with 2.5 GHz. Finally, the protein structure alignment browser (PSAB) appears, listing all successful protein structure alignment results with a Cα RMSD of less than ∼4 Å and at least 20 aligned residues. The latter restriction is imposed to avoid listing of meaningless alignment results. Each protein structure alignment result can be visualized in 3D and evaluated with regard to structure and sequence similarity. In the following section the functionality of the PSAB is demonstrated in detail for the selection of circular permuted protein structures from the GIS. The PSAB of the GIS web server consists of two parts (Figure 4).
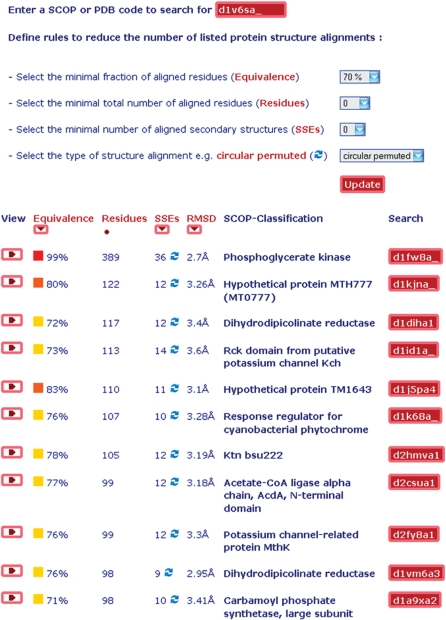
Figure 4.
Structure alignment browser (all-against-all) available at http://agknapp.chemie.fu-berlin.de/gplus. Top part: rules to search and reduce the number of listed protein structure alignments. Bottom part: sorted list of selected protein structure alignments with >70% of aligned residues <4 Å RMSD for the phosophoglycerate kinase with PDB id 1V6S, chain A relative to all proteins of the ASTRAL40 (SCOP 1.75) database of 10 444 protein structures.
The top part provides selection and filter criteria restricting the number of listed protein structure alignments. These are: minimum fraction of aligned residues with regard to the smaller of both protein structures (equivalence); minimum number of aligned residues (residues); and the minimum number of aligned SSEs. Furthermore, users may specify to list ‘circular permuted’ protein structure alignments only. A more detailed description of the mentioned selection criteria is given in section 2.1.
The bottom part of the structure alignment browser (all-against-all) (Figure 4) contains the list of all GIS protein structure alignments, which fulfill the beforehand specified criteria, giving PDB (18) and SCOP (27) id of the proteins. The list of protein structure alignments provides several features of the detected structural similarities such as fraction and number of aligned residues, number of aligned SSEs, and Cα RMSD. The list can subsequently be sorted according to these alignment features.
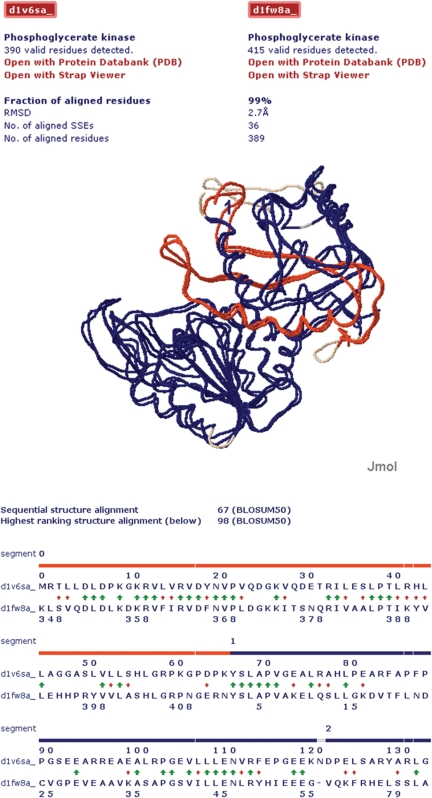
In the given example, the filter attribute ‘circular permuted’ was specified such that only protein structure alignments with exactly one break in the sequential order of assigned SSEs were listed. The protein structure alignments can be visualized with Jmol (http://www.jmol.org/) as shown in Figure 5 for the circular permuted protein structure alignment of 1V6S (29) and 1FW8 (30). Additionally, structure-based sequence alignments of the two proteins were generated. These were performed for both, the highest ranked structure alignment (sequential or non-sequential) and the highest ranked sequential structure alignment. The former is shown in detail (see Figure 5). The resulting net sequence similarities [BLOSUM50 scores (28)] are shown for both alignments just below the displayed structure in Figure 5. For the sequential structure-based sequence alignment, the BLOSUM50 score of 1V6S (29) and 1FW8 (30) was 67. In contrast, the non-sequential structure-based sequence alignment yields a BLOSUM50 score of 98. In addition to the high degree of structural similarity, the increase in sequence similarity going along with such a structure based non-sequential sequence alignment further indicates that the detected structurally similar protein pair is evolutionary related by circular permutation.
Figure 5.
Visualization of the circular permuted structure alignment result for the two proteins with PDB id 1V6S, chain A and 1FW8, chain A. Top part: protein structure alignment visualized with Jmol (see http://jmol.sourceforge.net for details). The aligned segments are shown in red and blue. Unaligned segments are shown in grey. Bottom part: illustration of the structure-based sequence alignment results evaluated with the BLOSUM50 substitution matrix (28). The color code is the same as in top part. The sequence break of the circular permutation (gap in blue band) has been detected between residues 66 and 67 in 1V6S, chain A. The green arrows indicate identical residue pairs and the red arrows residue pairs with a positive BLOSUM50 substitution score.
Usage from external software applications
We offer a web script (at http://agknapp.chemie.fu-berlin.de/gplus/addons/gis_info.php), which provides a list of similar protein structures and the corresponding alignment details in text-file format for a given PDB (18) or SCOP (27) id. The script can be integrated as a module in external programs specifying several Uniform Resource Locator (URL) command line parameters such as e.g. the PDB or SCOP id (id) and the total number of listed protein structures (n). Parameters are specified by adding a question mark ‘?’ to the given URL followed by the form ‘parameter name = parameter value’. If more than one URL parameter is specified, each has to be separated by an ampersand ‘&’.
CONCLUSION
The presented GIS is a comprehensive source for the analysis of structural relationships among protein structures. Users may compare and analyze (in sequential and non-sequential alignment mode) sequence and structure similarities for any given protein structure with the ASTRAL40 database. The provided service allows accessing the results in several ways i.e. using the internet pages or external software applications.
FUNDING
Funding for open access charge: Humboldt Universität zu Berlin.
Conflict of interest statement. None declared.
REFERENCES
- 1.Holm L, Park J. DaliLite workbench for protein structure comparison. Bioinformatics. 2000;6:566–567. doi: 10.1093/bioinformatics/16.6.566. [DOI] [PubMed] [Google Scholar]
- 2.Szustakowski JD, Weng Z. Protein structure alignment using a genetic algorithm. Proteins. 2000;38:428–440. doi: 10.1002/(sici)1097-0134(20000301)38:4<428::aid-prot8>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- 3.Shindyalov IN, Bourne PE. Protein structure alignment by incremental combinatorial extension (CE) of the optimal path. Protein Eng. 1998;11:739–747. doi: 10.1093/protein/11.9.739. [DOI] [PubMed] [Google Scholar]
- 4.Zhang Y, Skolnick J. TM-align: A protein structure alignment algorithm based on TM-score. Nucleic Acids Res. 2005;33:2302–2309. doi: 10.1093/nar/gki524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dror O, Benyamini H, Nussinov R, Wolfson HJ. MASS: multiple structural alignment by secondary structures. Bioinformatics. 2003;19:95–104. doi: 10.1093/bioinformatics/btg1012. [DOI] [PubMed] [Google Scholar]
- 6.Ilyin V, Abyzov A, Leslin C. Structural alignment of proteins by a novel TOPOFIT method, as a superimposition of common volumes at a topomax point. Protein Sci. 2004;13:1865–1874. doi: 10.1110/ps.04672604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bystroff Y. Non-sequential structure-based alignments reveal topology-independent core packing arrangements in proteins. Bioinformatics. 2005;7:1010–1019. doi: 10.1093/bioinformatics/bti128. [DOI] [PubMed] [Google Scholar]
- 8.Kolbeck B, May P, Schmidt-Goenner T, Steinke T, Knapp EW. Connectivity independent protein-structure alignment. BMC Bioinformatics. 2006;7:510–529. doi: 10.1186/1471-2105-7-510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bauer RA, Bourne PE, Formella A, Frömmel C, Gille C, Goede A, Guerler A, Hoppe A, Knapp EW, Pöschel T, et al. Superimposé: a 3D structural superposition server. Nucleic Acids Res. 2008;36:W47–W57. doi: 10.1093/nar/gkn285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shih ESC, Hwang MJ. Alternative alignments from comparison of protein structures. Proteins. 2004;59:519–527. doi: 10.1002/prot.20124. [DOI] [PubMed] [Google Scholar]
- 11.Chen L, Wu LY, Wang Y, Zhang S, Zhang XS. Revealing divergent evolution, identifying circular permutations and detecting active-sites by protein structure comparison. BMC Struct. Biol. 2006;6:18–31. doi: 10.1186/1472-6807-6-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guerler A, Knapp EW. Novel protein folds and their non-sequential structural analogs. Protein Sci. 2008;17:1374–1382. doi: 10.1110/ps.035469.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guerler A, Wang C, Knapp EW. Symmetric structures in the universe of protein folds. J. Chem. Inf. Model. 2009;49:2147–2151. doi: 10.1021/ci900185z. [DOI] [PubMed] [Google Scholar]
- 14.Schmidt-Goenner T, Guerler A, Kolbeck B, Knapp EW. Circular permuted proteins in the universe of protein folds. Proteins: Structure, Function, and Bioinformatics. 2009;78:1618–1630. doi: 10.1002/prot.22678. [DOI] [PubMed] [Google Scholar]
- 15.Guerler A, Knapp EW. Strategies of nonsequential structure alignments. Genome Informatics. 2009;22:21–29. [PubMed] [Google Scholar]
- 16.Guerler A, Knapp EW. Evaluation of sequence alignments of distantly related sequence pairs with respect to structural similarity. Genome Informatics. 2007;18:183–191. [PubMed] [Google Scholar]
- 17.Gille C, Frömmel C. STRAP: editor for STRuctural Alignments of Proteins. Bioinformatics. 2000;17:377–378. doi: 10.1093/bioinformatics/17.4.377. [DOI] [PubMed] [Google Scholar]
- 18.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Han BW, Bingman CA, Mahnke DK, Bannen RM, Bednarek SY, Sabina RL, Phillips GN. Membrane association, mechanism of action, and structure of Arabidopsis embryonic factor 1 (FAC1) J. Biol. Chem. 2006;281:14939–14947. doi: 10.1074/jbc.M513009200. [DOI] [PubMed] [Google Scholar]
- 20.Benini S, Rypniewski WR, Wilson KS, Ciurli S, Mangani S. Structure-based rationalization of urease inhibition by phosphate: novel insights into the enzyme mechanism. J. Biol. Inorg. Chem. 2001;6:778–790. doi: 10.1007/s007750100254. [DOI] [PubMed] [Google Scholar]
- 21.Hasegawa H, Holm L. Advances and pitfalls of protein structural alignment. Curr. Opin. Struct. Biol. 2009;19:341–348. doi: 10.1016/j.sbi.2009.04.003. [DOI] [PubMed] [Google Scholar]
- 22.Orengo CA, Taylor WR. SSAP: sequential structure alignment program for protein structure comparison. Methods Enzymol. 1996;266:617–635. doi: 10.1016/s0076-6879(96)66038-8. [DOI] [PubMed] [Google Scholar]
- 23.Zemla A. LGA-a method for finding 3D similarities in protein structures. Nucleic Acids Res. 2003;31:3370–3374. doi: 10.1093/nar/gkg571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Standley DM, Toh H, Nakamura H. GASH: an improved algorithm for maximizing the number of equivalent residues between two protein structures. BMC Bioinform. 2005;6:22–40. doi: 10.1186/1471-2105-6-221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Csaba G, Birzele F, Zimmer R. Protein structure alignment considering phenotypic plasticity. Bioinformatics. 2008;24:i98–i104. doi: 10.1093/bioinformatics/btn271. [DOI] [PubMed] [Google Scholar]
- 26.DeLano WL. The PyMOL Molecular Graphics System. 2002 http://www.pymol.org/ (17 April, 2010, date last accessed) [Google Scholar]
- 27.Murzin AG, Brenner SE, Hubbard T, Chothia C. SCOP. J. Mol. Biol. 1995;247:536–540. doi: 10.1006/jmbi.1995.0159. [DOI] [PubMed] [Google Scholar]
- 28.Henikoff S, Henikoff JG. Amino acid substitution matrices from protein blocks. Proc. Natl Acad. Sci. USA. 1992;89:10915–10919. doi: 10.1073/pnas.89.22.10915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mizutani H, Kunishima N. Crystal structure of phosphoglycerate kinase from thermus thermophilus HB8. In press. [Google Scholar]
- 30.Tougard P, Bizebard T, Ritco-Vonsovici M, Minard P, Desmadril M. Structure of a circularly permuted phosphoglycerate kinase. Acta Crystallogr. D. 2002;58:2018–2023. doi: 10.1107/s0907444902015548. [DOI] [PubMed] [Google Scholar]