Abstract
Genome-wide association study (GWAS) is nowadays widely used to identify genes involved in human complex disease. The standard GWAS analysis examines SNPs/genes independently and identifies only a number of the most significant SNPs. It ignores the combined effect of weaker SNPs/genes, which leads to difficulties to explore biological function and mechanism from a systems point of view. Although gene set enrichment analysis (GSEA) has been introduced to GWAS to overcome these limitations by identifying the correlation between pathways/gene sets and traits, the heavy dependence on genotype data, which is not easily available for most published GWAS investigations, has led to limited application of it. In order to perform GSEA on a simple list of GWAS SNP P-values, we implemented GSEA by using SNP label permutation. We further improved GSEA (i-GSEA) by focusing on pathways/gene sets with high proportion of significant genes. To provide researchers an open platform to analyze GWAS data, we developed the i-GSEA4GWAS (improved GSEA for GWAS) web server. i-GSEA4GWAS implements the i-GSEA approach and aims to provide new insights in complex disease studies. i-GSEA4GWAS is freely available at http://gsea4gwas.psych.ac.cn/.
INTRODUCTION
Genome-wide association study (GWAS) has become a popular approach by utilizing genome-wide genotyping array to map susceptibility effects through examining associations between SNP genotype frequency and traits (1,2). The standard GWAS analysis focuses on single SNP/gene and identifies only a number of the most significant SNPs that account for only a small proportion of the genetic variants and offers limited understanding of complex diseases (3). It also ignores the combined SNP/gene effects representing interactions of multiple genetic factors of complex disease, which lead to difficulties in exploring biological function and mechanism from a systems point of view.
To overcome these limitations, the principle of gene set analysis (GSA), which was recently developed to facilitate handling of genome-wide expression data (4), has been introduced to GWAS. GSA tests the correlation between gene set, which is defined as genes involved in the same pathway, and trait, resting on the assumption that genes belonging to a disease-related pathway/gene set trend to reveal concordant signals. Recently, several implementations or application programs using the GSA principle have been developed (3,5–14). An interesting implementation is the gene set enrichment analysis (GSEA) which technically evaluates whether the distribution of genes sharing a biochemical or cellular function is different from the distribution of a ranked genome-wide gene list by computing Kolmogorov–Smirnov like statistics (3,15). Successful applications of GSEA in GWAS include the study of Parkinson’s disease (3), Crohn’s disease (11) and the study of host control for HIV-1 (9).
However, GSEA usually depends on genotype data, which is not easily available for most published GWAS investigations. This leads to a limited application of it, especially application in the secondary analysis of published GWASs to best interpret the remaining long list of GWAS data from a systems point of view. In order to perform GSEA on easily available GWAS data, mainly SNP P-values, we implemented GSEA by using SNP label permutation instead of phenotype label permutation to analyze P-values. We further improved the GSEA (i-GSEA) by focusing on pathways/gene sets with high proportions of significant genes instead of relying only on the total significance coming from either a few or many significant genes. Our study shows that i-GSEA has the improved sensitivity to identify pathways/gene sets representing combined effects of possibly modest SNPs/genes.
There have been more than 480 GWAS publications covering a broad range of human diseases available by the end of January 2010 [see http://www.genome.gov/gwastudies/ and (16)] and the number keeps increasing. In order to implement the i-GSEA approach and provide researchers with an open platform to analyze the easily available GWAS data from both publications and self experiments, we developed the i-GSEA4GWAS (improved GSEA for GWAS) web server. i-GSEA4GWAS aims to identify novel pathways/gene sets correlated with given traits and provide new insights into complex disease study, especially in complementation with standard single SNP/gene based GWAS analysis.
Overview of the i-GSEA algorithm
The i-GSEA4GWAS web server implements i-GSEA to help researchers explore GWAS data efficiently. i-GSEA is an application and extension of GSEA. For GSEA, it runs the following three key procedures. (i) The max statistics or −log(P-value) of closely spaced SNPs in a gene is used to represent the gene; then, the ranked gene list with corresponding representing values is utilized to calculate each gene set’s enrichment score (ES), i.e. a Kolmogorov–Smirnov like statistics with weight 1, which reflects the trend that genes of a gene set tend to be located at the top of the entire ranked genome-wide gene list. (ii) The phenotype label permutation (to break the association between genotype and phenotype) and a straightforward normalization are performed to generate the distribution of the ES and correct gene variation (i.e. different genes with different number of SNPs mapped will result in identification of gene sets containing genes with more SNPs mapped, instead of genes with functional correlation) and gene set variation (i.e. different gene sets contain different number of genes) (3). (iii) Based on all the distributions of ESs generated by permutation, false discovery rate (FDR) is used for multiple testing correction (17).
In i-GSEA, we implement SNP label permutation instead of phenotype label permutation to analyze SNP P-values and to correct gene and gene set variation and multiply k/K to the ES to get the significance proportion based enrichment score (SPES), where k is the proportion of significant genes of the gene set and K is the proportion of significant genes of the total genes in the GWAS. Here, significant genes are defined as the genes mapped with at least one of the top 5% of all SNPs. Instead of ES which focuses on the total significance coming from either a few or many significant genes, SPES emphasizes on total significance coming from high proportion of significant genes. So, i-GSEA trends to pick up pathways/gene sets including a high proportion of significant genes and is more appropriate for study of the combined effects of possibly modest SNPs/genes in complex disease. This ensures i-GSEA the improved sensitivity.
Description of the i-GSAE4GWAS web server
System development, configuration and system overview
i-GSEA4GWAS is written in Java and JSP. The program is running in a high-end computing cluster with six dedicated Intel XEON Quad-Core CPUs at 2.0 GHz and two dedicated Intel XEON Quad-Core CPUs at 3.0 GHz (representing a total of 32 cores) with a total of 96 GB of memory.
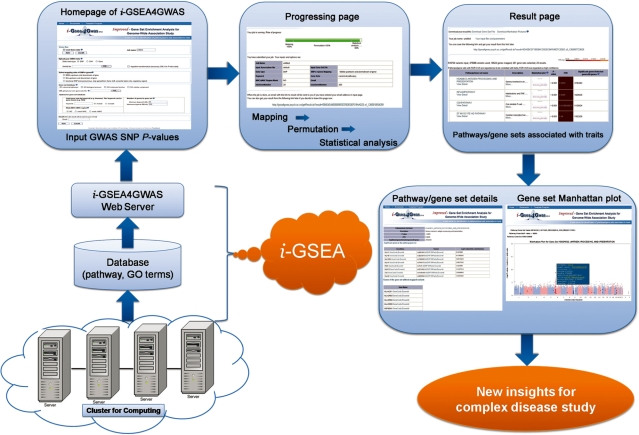
i-GSEA4GWAS is freely available and there is NO registration required. We also provide a template program for users to access our web server through command lines instead of web browser. Figure 1 shows the system architecture and overview of our i-GSEA4GWAS web server.
Figure 1.
The system architecture and overview of i-GSEA4GWAS web server.
Input data
The input data of i-GWAS are SNP data, mainly P-values. The server will transform the P-value to −log(P-value). Some other values of association test like odds ratio and statistics are also accepted. Here, we define all these input values as ‘association value’. The detailed format of input is a SNP list with two columns, namely SNP identifier and its corresponding association value. A gene list with gene HUGO symbol (http://www.genenames.org/) (18) and along with corresponding association value is also accepted but not recommended due to the limitation of gene variation as described above.
The collection of pathways/gene sets utilized in i-GSEA4GWAS are extracted and curated from MSigDB v2.5 (http://www.broadinstitute.org/gsea/msigdb/) (15) to ensure comprehensiveness and high quality. It includes gene sets denoting canonical pathways integrated from a variety of online resources like KEGG (19) and BioCarta (http://www.biocarta.com/), and GO (gene ontology) terms with high confidence (20). Additionally, i-GSEA4GWAS supports customized gene sets, i.e. users can upload their own gene sets. This provides the opportunity that the knowledge database (gene sets) can always be expanded, and makes it easy for users to test their hypothesis within their research interest.
Running procedure
First, our server maps SNPs to the genome-wide genes. i-GSEA4GWAS contains an optional multiple-level broad-to-narrow SNPs->genes mapping system, which helps users either fully explore genome-wide SNP data (by choosing ‘500-kb upstream and downstream of gene’) or narrow down the research scope by only focusing on functional SNPs (by choosing ‘functional SNP’). The SNPs->genes mapping is established based on SNP and gene annotations from the Ensembl BioMart database (Release 56–15 September 2009, http://www.ensembl.org/biomart/martview) (21). Next, the program uses the list of SNP-mapped genes to filter the collection of pathways/gene sets to obtain candidate pathways/gene sets. Additional filtering includes a keyword search to define the function of gene sets, to mask major histocompatibility complex/extended major histocompatibility complex (MHC/xMHC) region (22) to only explore the non-MHC/xMHC region (23), and to restrain gene set size to avoid overly narrow or overly broad functional categories and too large a search space, with the default parameter of minimum 20 and maximum 200 (3,9). Then i-GSEA4GWAS performs i-GSEA to identify pathways/gene sets correlated with traits. When the job is running, the progress of the three key steps (mapping, permutation and statistical analysis) will be shown in the progress bar.
Output and examples
The pathways/gene sets, with FDR < 0.25 (the threshold of FDR < 0.25 denotes the confidence of ‘possible’ or ‘hypothesis’, while the threshold of FDR < 0.05 is regarded as ‘high confidence’ or ‘with statistical significance’), are displayed in a tabular form with links to detailed text information of pathways/gene sets, genes and SNPs. One characteristic feature of i-GSEA4GWAS output is the Manhattan plot of gene set, which uses the Manhattan plot of GWAS as background, and highlights the results of association test for a given pathway/gene set. It helps users to compare the association results of the given pathway/gene set with the genome-scale data graphically. The result page also contains the download URL, from where all the results, both texts and figures, can be downloaded.
The first example data is SNP P-value from the GWAS of HIV-1 host control investigation, which studied the quantitative trait—HIV-1 viral load set point and used Illumina HumanHap550 BeadChip (24). The input includes 518 754 SNPs with corresponding P-values. There are 479 086 SNPs mapped to 16 524 genes by using the default ‘500 kb upstream and downstream of gene’. 251 gene sets were selected by using ‘canonical pathways’, and using the default gene set size ranges from 20 through 200. A total of 23 pathways/gene sets were identified to be possibly correlated with the trait (FDR < 0.25). Among the 23 pathways, five pathways namely ‘antigen processing and presentation’, ‘inflampathway’, ‘cskpathway’, ‘ribosome’ and ‘st myocyte ad pathway’ have high confidence (FDR < 0.05). The pathways ‘antigen processing and presentation’ (FDR = 0.009), ‘inflampathway’ (FDR = 0.010), and ‘cskpathway’ (FDR = 0.011) have been replicated in our follow-up work of the initial GWAS by either the top hits or using GSEA (9). Both the pathways ‘ribosome’ and ‘st myocyte ad pathway’ were only identified by i-GSEA (FDR < 0.05) and were not replicated by the follow-up GWAS and GSEA analysis. However, both pathways have references to support their relevance to HIV. For pathway ‘ribosome’ (FDR = 0.047), it has been known that the ribosomal frame shifting is a kind of gene expression mechanism of several RNA viruses (including HIV-1 and SARS-CoV) to express replicase enzymes and a couple of investigations have been carried out on this topic (25–27). The pathway ‘st myocyte ad pathway’ (FDR = 0.040) contains a variety of adrenergic receptors that induce subtype-specific signaling effects. The top gene with the smallest P-value in this gene set, ITPR1 (inositol 1,4,5-triphosphate receptor, type 1), has been reported to interact with genes in HIV. For example HIV-1 Nef interacts with ITPR1 to trigger the activation of plasma membrane calcium influx channels (28); HIV-1 Tat induces release of calcium from ITPR1-regulated stores in neurons and astrocytes, an effect that plays an important role in Tat-induced TNF-α production (29).
The second example data are SNP P-values of the GWAS of bipolar disorder, which is a case/control study and utilized the GeneChip 500K Mapping Array Set (Affymetrix chip) (2). i-GSEA4GWAS identified two pathways/gene sets (FDR < 0.05) by using the default ‘500 kb upstream and downstream of gene’ SNPs- > genes mapping rule, searching ‘canonical pathways’ and GO terms, and masking the xMHC region. One pathway is ‘glutathione metabolism’ (FDR = 0.036), which was also reported by Holmans et al.(30). Reducing glutathione (GSH) level will lead to an increased potential for cellular oxidative stress, which has been implicated in the pathology of bipolar disorder, and recently some studies have been performed on animal models (31,32). The other one is ‘cysteine type endopeptidase activity’ (FDR = 0.041), which might be a novel functional biological mechanism related to bipolar disorder and worthy of further study. There is no pathway/gene set found with FDR < 0.05 or FDR < 0.25 by using GSEA with the same parameters.
DISCUSSION
Our web server implements i-GSEA for GWAS data analysis to explore the biological function and mechanism from a systems point of view. It is efficient not only for in-house GWASs with available genotype data, but also for secondary data mining of published GWASs, where genotype data are not readily available. i-GSEA4GWAS aims to help researchers take the best advantage of the high-cost GWAS data to look for new biological insights worthy of further confirmation. This represents the major potential for i-GSEA4GWAS. The improved i-GSEA approach, the optimal multiple-level SNPs->genes mapping system, the comprehensive pathway/gene set database with options of customization, as well as the consideration of MHC/xMHC region distinguish i-GSEA4GWAS from other programs. It should be noted that i-GSEA4GWAS does not take into account the linkage disequilibrium (LD) patterns from SNP arrays and would not prune the set of SNPs for LD since these can only be done when genotype data is available. So, users are recommended to input SNPs not in LD (say r2 < 0.2) to reduce the possibility of biased results due to LD patterns from SNP arrays. On the other hand, as the name of the web server defines, i-GSEA4GWAS is only applicable to whole-genome SNP arrays.
i-GSEA employs SNP label permutation to correct gene variation to reduce the bias due to different genes with different number of mapped SNPs, which is ignored by the non-randomization approach such as the segmentation test (33) as implemented in the GeSBAP web server (10). This correction ensures to identify gene sets consisting of non-random high-association genes with biological plausibility instead of random high-association genes with large numbers of mapped SNPs. Furthermore, the key issue of i-GSEA, focusing on pathways/gene sets with high proportion of significant genes to detect combinations of modest effects, greatly improves sensitivity. Let n denote the number of pathways with FDR < 0.05 and m represent the number of pathways with both FDR < 0.05 and references to support, our comparison study between i-GSEA and GSEA shows, in the HIV-1 host control and bipolar disorder study as described above, n = 7 and m = 6 for i-GSEA, while n = 2 and m = 2 for GSEA. Further comparison by using the other six Wellcome Trust Case Control Consortium GWASs except bipolar disorder (2) obtained the result that n = 33 and m = 14 for i-GSEA, while n = 2, m = 1 for GSEA. In addition, the pathways identified (FDR < 0.05) by i-GSEA include all the pathways identified by GSEA in all the above comparisons. These show that i-GSEA has improved sensitivity in comparison to GSEA. Our data examples also show that i-GSEA4GWAS obtains very meaningful results with different GWAS designs (quantitative trait and case/control) and different genotyping platforms (Illumina and Affymetrix).
There are always conflicts between the amount and the quality of the gene sets utilized for computation. Using a large amount of gene sets, such as all GO terms, will introduce large background and noise due to the large number and low confidence of some gene sets, while only using a few gene sets will lose information. Moreover, the knowledge of gene sets is keeping increasing. To overcome this, we chose a balanced strategy, using a curated pathway/gene set database with consideration of both comprehensiveness and high quality, and allow researchers to upload their customized gene sets to make sure the gene sets of research focus will be well represented. This ensures that i-GSEA4GWAS contains a high-quality and reasonable search space. The i-GSEA4GWAS will be regularly updated to ensure the most up-to-date searching database and annotations. i-GSEA4GWAS also integrates the curated copy number variations (CNVs) from the Database of Genomic Variants (http://projects.tcag.ca/variation/) (34). In the future, the CNV functional module will be further extended to include the CNV probes of Illumina, Affymetrix and more genome-wide genotyping arrays.
In summary, the i-GSEA4GWAS web server provides researchers an efficient open platform for GWAS analysis, helping further interpret the SNP P-values from hundreds of available GWASs and future GWASs to provide new insights into disease study.
FUNDING
Project for Young Scientists Fund, Institute of Psychology, Chinese Academy of Sciences (O9CX115011); National Natural Science Foundation of China (NSFC) (30700441); and the Beijing New Star Project, Beijing Municipal Science & Technology Commission Foundation (2007A082). Funding for open access charge: O9CX115011.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We thank Dr Lei Kong and Dr Ge Gao from Peking University for their kind help on the system configurations. We thank all our colleagues and friends in Chinese Academy of Sciences, who helped test the web server and provided us valuable suggestions. We thank the anonymous reviewers for their helpful comments and suggestions.
REFERENCES
- 1.McCarthy MI, Abecasis GR, Cardon LR, Goldstein DB, Little J, Ioannidis JP, Hirschhorn JN. Genome-wide association studies for complex traits: consensus, uncertainty and challenges. Nat. Rev. Genet. 2008;9:356–369. doi: 10.1038/nrg2344. [DOI] [PubMed] [Google Scholar]
- 2.Wellcome Trust Case Control Consortium. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang K, Li M, Bucan M. Pathway-Based Approaches for Analysis of Genomewide Association Studies. Am. J. Hum. Genet. 2007;81:1278–1283. doi: 10.1086/522374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nam D, Kim SY. Gene-set approach for expression pattern analysis. Brief. Bioinform. 2008;9:189–197. doi: 10.1093/bib/bbn001. [DOI] [PubMed] [Google Scholar]
- 5.Holden M, Deng S, Wojnowski L, Kulle B. GSEA-SNP: applying gene set enrichment analysis to SNP data from genome-wide association studies. Bioinformatics. 2008;24:2784–2785. doi: 10.1093/bioinformatics/btn516. [DOI] [PubMed] [Google Scholar]
- 6.Torkamani A, Topol EJ, Schork NJ. Pathway analysis of seven common diseases assessed by genome-wide association. Genomics. 2008;92:265–272. doi: 10.1016/j.ygeno.2008.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Askland K, Read C, Moore J. Pathways-based analyses of whole-genome association study data in bipolar disorder reveal genes mediating ion channel activity and synaptic neurotransmission. Hum. Genet. 2009;125:63–79. doi: 10.1007/s00439-008-0600-y. [DOI] [PubMed] [Google Scholar]
- 8.Chen L, Zhang L, Zhao Y, Xu L, Shang Y, Wang Q, Li W, Wang H, Li X. Prioritizing risk pathways: a novel association approach to searching for disease pathways fusing SNPs and pathways. Bioinformatics. 2009;25:237–242. doi: 10.1093/bioinformatics/btn613. [DOI] [PubMed] [Google Scholar]
- 9.Fellay J, Ge D, Shianna KV, Colombo S, Ledergerber B, Cirulli ET, Urban TJ, Zhang K, Gumbs CE, Smith JP, et al. Common genetic variation and the control of HIV-1 in humans. PLoS Genet. 2009;5:e1000791. doi: 10.1371/journal.pgen.1000791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Medina I, Montaner D, Bonifaci N, Pujana MA, Carbonell J, Tarraga J, Al-Shahrour F, Dopazo J. Gene set-based analysis of polymorphisms: finding pathways or biological processes associated to traits in genome-wide association studies. Nucleic Acids Res. 2009;37:W340–W344. doi: 10.1093/nar/gkp481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang K, Zhang H, Kugathasan S, Annese V, Bradfield JP, Russell RK, Sleiman PM, Imielinski M, Glessner J, Hou C, et al. Diverse genome-wide association studies associate the IL12/IL23 pathway with Crohn Disease. Am. J. Hum. Genet. 2009;84:399–405. doi: 10.1016/j.ajhg.2009.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yu K, Li Q, Bergen AW, Pfeiffer RM, Rosenberg PS, Caporaso N, Kraft P, Chatterjee N. Pathway analysis by adaptive combination of P-values. Genet. Epidemiol. 2009;33:700–709. doi: 10.1002/gepi.20422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Peng G, Luo L, Siu H, Zhu Y, Hu P, Hong S, Zhao J, Zhou X, Reveille JD, Jin L, et al. Gene and pathway-based second-wave analysis of genome-wide association studies. Eur. J. Hum. Genet. 2010;18:111–117. doi: 10.1038/ejhg.2009.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cantor RM, Lange K, Sinsheimer JS. Prioritizing GWAS results: A review of statistical methods and recommendations for their application. Am. J. Hum. Genet. 2010;86:6–22. doi: 10.1016/j.ajhg.2009.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl Acad. Sci. USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hindorff LA, Sethupathy P, Junkins HA, Ramos EM, Mehta JP, Collins FS, Manolio TA. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc. Natl Acad. Sci. USA. 2009;106:9362–9367. doi: 10.1073/pnas.0903103106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Reiner A, Yekutieli D, Benjamini Y. Identifying differentially expressed genes using false discovery rate controlling procedures. Bioinformatics. 2003;19:368–375. doi: 10.1093/bioinformatics/btf877. [DOI] [PubMed] [Google Scholar]
- 18.Eyre TA, Ducluzeau F, Sneddon TP, Povey S, Bruford EA, Lush MJ. The HUGO Gene Nomenclature Database, 2006 updates. Nucleic Acids Res. 2006;34:D319–D321. doi: 10.1093/nar/gkj147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ogata H, Goto S, Sato K, Fujibuchi W, Bono H, Kanehisa M. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 1999;27:29–34. doi: 10.1093/nar/27.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hubbard TJ, Aken BL, Ayling S, Ballester B, Beal K, Bragin E, Brent S, Chen Y, Clapham P, Clarke L, et al. Ensembl 2009. Nucleic Acids Res. 2009;37:D690–D697. doi: 10.1093/nar/gkn828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Horton R, Wilming L, Rand V, Lovering RC, Bruford EA, Khodiyar VK, Lush MJ, Povey S, Talbot C.C., Jr, Wright MW, et al. Gene map of the extended human MHC. Nat. Rev. Genet. 2004;5:889–899. doi: 10.1038/nrg1489. [DOI] [PubMed] [Google Scholar]
- 23.Warrens A, Lechler R. HLA in Health and Disease. San Diego, CA: Academic Press; 1999. [Google Scholar]
- 24.Fellay J, Shianna KV, Ge D, Colombo S, Ledergerber B, Weale M, Zhang K, Gumbs C, Castagna A, Cossarizza A, et al. A whole-genome association study of major determinants for host control of HIV-1. Science. 2007;317:944–947. doi: 10.1126/science.1143767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mazauric MH, Seol Y, Yoshizawa S, Visscher K, Fourmy D. Interaction of the HIV-1 frameshift signal with the ribosome. Nucleic Acids Res. 2009;37:7654–7664. doi: 10.1093/nar/gkp779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gendron K, Charbonneau J, Dulude D, Heveker N, Ferbeyre G, Brakier-Gingras L. The presence of the TAR RNA structure alters the programmed -1 ribosomal frameshift efficiency of the human immunodeficiency virus type 1 (HIV-1) by modifying the rate of translation initiation. Nucleic Acids Res. 2008;36:30–40. doi: 10.1093/nar/gkm906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brierley I, Dos Ramos FJ. Programmed ribosomal frameshifting in HIV-1 and the SARS-CoV. Virus Res. 2006;119:29–42. doi: 10.1016/j.virusres.2005.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Manninen A, Saksela K. HIV-1 Nef interacts with inositol trisphosphate receptor to activate calcium signaling in T cells. J. Exp. Med. 2002;195:1023–1032. doi: 10.1084/jem.20012039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mayne M, Holden CP, Nath A, Geiger JD. Release of calcium from inositol 1,4,5-trisphosphate receptor-regulated stores by HIV-1 Tat regulates TNF-alpha production in human macrophages. J. Immunol. 2000;164:6538–6542. doi: 10.4049/jimmunol.164.12.6538. [DOI] [PubMed] [Google Scholar]
- 30.Holmans P, Green EK, Pahwa JS, Ferreira MA, Purcell SM, Sklar P, Owen MJ, O'Donovan MC, Craddock N. Gene ontology analysis of GWA study data sets provides insights into the biology of bipolar disorder. Am. J. Hum. Genet. 2009;85:13–24. doi: 10.1016/j.ajhg.2009.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dean OM, van den Buuse M, Bush AI, Copolov DL, Ng F, Dodd S, Berk M. A role for glutathione in the pathophysiology of bipolar disorder and schizophrenia? Animal models and relevance to clinical practice. Curr. Med. Chem. 2009;16:2965–2976. doi: 10.2174/092986709788803060. [DOI] [PubMed] [Google Scholar]
- 32.Dean O, Bush AI, Berk M, Copolov DL, van den Buuse M. Glutathione depletion in the brain disrupts short-term spatial memory in the Y-maze in rats and mice. Behav. Brain Res. 2009;198:258–262. doi: 10.1016/j.bbr.2008.11.017. [DOI] [PubMed] [Google Scholar]
- 33.Al-Shahrour F, Arbiza L, Dopazo H, Huerta-Cepas J, Minguez P, Montaner D, Dopazo J. From genes to functional classes in the study of biological systems. BMC Bioinformatics. 2007;8:114. doi: 10.1186/1471-2105-8-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Iafrate AJ, Feuk L, Rivera MN, Listewnik ML, Donahoe PK, Qi Y, Scherer SW, Lee C. Detection of large-scale variation in the human genome. Nat. Genet. 2004;36:949–951. doi: 10.1038/ng1416. [DOI] [PubMed] [Google Scholar]