Abstract
Transposable elements (TE) exist in the genomes of nearly all eukaryotes. TE mobilization through ‘cut-and-paste’ or ‘copy-and-paste’ mechanisms causes their insertions into other repetitive sequences, gene loci and other DNA. An insertion of a TE commonly creates a unique TE junction in the genome. TE junctions are also randomly distributed along chromosomes and therefore useful for genome-wide marker development. Several TE-based marker systems have been developed and applied to genetic diversity assays, and to genetic and physical mapping. A software tool ‘RJPrimers’ reported here allows for accurate identification of unique repeat junctions using BLASTN against annotated repeat databases and a repeat junction finding algorithm, and then for fully automated high-throughput repeat junction-based primer design using Primer3 and BatchPrimer3. The software was tested using the rice genome and genomic sequences of Aegilops tauschii. Over 90% of repeat junction primers designed by RJPrimers were unique. At least one RJM marker per 10 Kb sequence of A. tauschii was expected with an estimate of over 0.45 million such markers in a genome of 4.02 Gb, providing an almost unlimited source of molecular markers for mapping large and complex genomes. A web-based server and a command line-based pipeline for RJPrimers are both available at http://wheat.pw.usda.gov/demos/RJPrimers/.
INTRODUCTION
Transposable elements (TE) make up large proportions of many eukaryotic genomes. For example, they represent ∼35% of the rice genome (1), and ∼90% of hexaploid wheat genome (2) and significantly contribute to the size, organization and evolution of plant genomes. Based on the mechanism of transposition, TEs are classified into two major classes on the basis of the mechanism of transposition (3): Class I retrotransposons which transpose through ‘copy-and-paste’ mechanism, and Class II DNA transposons which transpose through ‘cut-and-paste’ mechanism. Their frequent amplification and transposition causes random insertion of TEs into other TE, gene or other DNA sequences, creating specific junctions between the newly inserted TE and surrounding sequences. Those TE or repeat junctions are commonly unique and genome-specific. They can be therefore treated as single copy markers in the genome (4,5). The genome specificity of TE junction-based markers makes them particularly useful for mapping of polyploid species including many important crops, such as wheat and cotton. Because repeat junctions are also abundant and randomly distributed along chromosomes (5), they have a great potential in development of genome-wide molecular markers for high-throughput mapping and diversity studies in large and complex genomes (5,6).
Several TE junction-based molecular marker systems have been developed and applied to genetic diversity assessments, and to genetic and physical mapping. These systems include sequence-specific amplification polymorphism (SSAP) (7), retrotransposon-microsatellite amplified polymorphism (REMAP) (8), retrotransposon-based insertion polymorphism (RBIP) (9), inter-retrotransposon amplified polymorphism (IRAP) (8), repeat junction marker (RJM) (5), repeat junction–junction marker (RJJM) (10) and insertion-site-based polymorphism (ISBP) (4,11). All those marker systems are based on the idea that TE junctions are unique. In some cases, the junctions are between two TEs of the same or different types (ISBP, IRAP, RJM, RJJM). In other cases, the junction could be between a TE and another sequence feature like a specific restriction site (SSAP), a microsatellite (REMAP) or a piece of unique sequence (RBIP). Hence, discovery of repeat junctions is a prerequisite to all those marker development systems.
Two possible approaches are available for repeat DNA discovery: de novo discovery and known repeat database based homology searching. Several software tools have been developed for detecting unknown long-terminal repeat (LTR) retrotransposons, such as LTR_STRUC (12) and LTR_FINDER (13), but those tools work for intact elements with well-conserved structural features. RECON is another tool for de novo discovery and grouping of unknown TE families (14). None of those tools are suitable for repeat junction identification in short sequences, such as individual Sanger shotgun sequences, bacterial artificial chromosome (BAC) end sequences, or reads from next generation sequencing technologies, because they usually do not cover complete conserved features of TEs. Although RepeatMasker (http://www.repeatmasker.org/) is the best program for database-based repeat DNA screening and masking, it is not designed to detect precise repeat junction sites. The junctions masked using RepeatMasker are not necessarily true junctions. This is because eukaryotic genomes contain large amounts of ancient, highly degenerated TEs and RepeatMasker fails to detect some of these ‘distant homologs’ of known TE families. JunctionViewer (15) is a most recently published software tool to identify and differentiate closely related centromere repeats and repeat junctions with graphical displays, but it’s not aimed to unique junction discovery and junction primer design.
A software tool for accurate identification of repeat junctions and automated PCR primer design will be useful for developing repeat junction-based marker systems for mapping of large and complex genomes. Here, we present a high-throughput, fully automated computational tool, RJPrimers, which employs a BLASTN search and a newly developed repeat junction finding algorithm to identify repeat junctions and then designs RJM primers. In addition, several other TE junction-based primer design strategies, including RJJM, ISBP, RBIP and IRAP, also are implemented in this tool. The abundance and uniqueness of TE junction-based markers designed by RJPrimers were examined in the rice genome (∼400 Mb) and in the large and complex A. tauschii genome (∼4.02 Gb), a diploid ancestor of bread wheat (Triticum aestivum).
SOFTWARE DEVELOPMENT
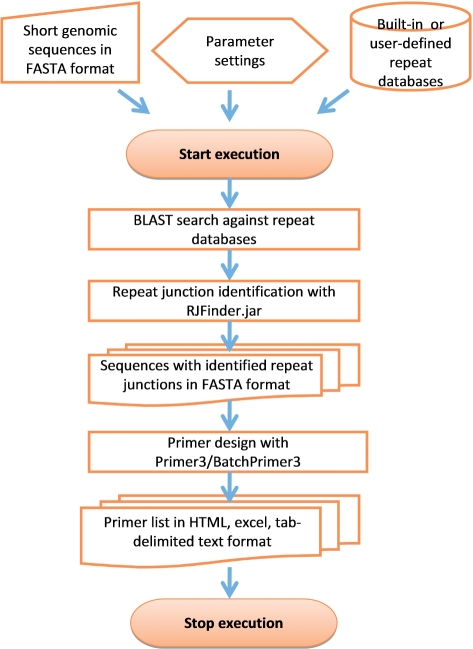
RJPrimers has been implemented in two versions, a web server and a command line-based pipeline equivalent. The web server acts as a convenient and easy-to-use tool for a limited number of DNA sequences, while the pipeline can be used for any amount of input data without memory limitation. However, both versions execute the same algorithm which consists of two dedicated steps, repeat junction identification and primer design. The procedures for designing and implementing RJPrimers software are illustrated in Figure 1.
Figure 1.
Workflow chart of RJPrimers.
Repeat junction identification
Repeat junction is identified by performing a homology search of the query DNA sequence as input in fasta format against known repeat databases. Several major repeat libraries for plant and animal genomes have been compiled from multiple public resources, such as RepBase (http://www.girinst.org/repbase/index.html), MIPS-REdat (16), TIGR plant repeat databases (17), maize TE database (maize TEDB, http://maizetedb.org/∼maize/) and Triticeae repeat database (TREP) (http://wheat.pw.usda.gov/ITMI/Repeats/). RepBase is a well organized and annotated plant and animal repeat database, and is a default library for the RepeatMasker program. MIPS-REdat, was compiled from several major plant repeat libraries using MIPS repeat element catalog (mips-REcat), a systematic hierarchical tree structure of repeat classifications. All repeat databases have inconsistent classification terms and cannot be directly used to clearly distinguish repeat junction types. To unify the repeat classifications with a controlled vocabulary, we adopted the unified classification system proposed by Wicker et al. (3), which includes levels of class, subclass, order, superfamily, family and subfamily. A Perl script was written to recompile RepBase, MIPS-REdat, TIGR plant repeat databases, maize TEDB and TREP in a specific format for RJPrimers. The recompiled repeat databases can be downloaded together with RJPrimers. In addition, user-defined databases can be also easily integrated to the command line based pipeline.
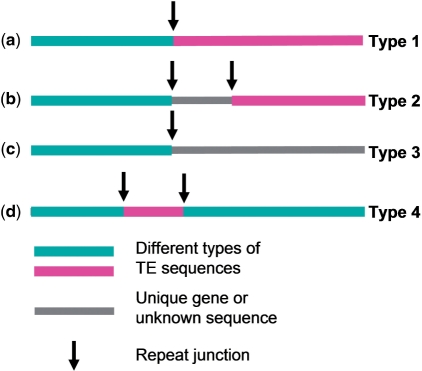
Structures of repeat junctions can be generally grouped into four basic types according to the sequence annotations at each side of the junction (Figure 2). To classify the junction type for a short sequence, a BLASTN search against a selected repeat database is performed. The BLASTN results are parsed and annotated for repeat junctions. A Java program ‘RJFinder’ was developed to take a sequence file as input and a repeat junction-containing sequence file as output. For a TE junction to be accurately detected, one region of the query sequence has to match at least one known TE sequence in the blast search. The second region can have a match to another known TE sequence, or can have no match. Based on our observation, a top hit with an E-value ≤1E−50 for the first match and a hit E-value ≤ 1E−5 for the second match are reasonable values for effective detection of ‘distant homologs’ of known TE families to classify repeat junction types. Therefore, these values are used in the default setting in the web program. The E-values from BLAST search were only used in identification of repeat junctions in the analysis.
Figure 2.
Repeat junction types in a short sequence. (a) Type 1: a repeat junction between two different TEs. (b) Type 2: two repeat junctions involving two different TE and an unknown or gene sequence. (c) Type 3: a repeat junction with a TE in one side and a fragment of an unknown or gene sequence on the other side. (d) Type 4: a nested repeat junction caused by a TE inserting into another TE.
A repeat junction is considered truly positive or unique if it meets any one of the following criteria: (i) Both sides of a potential junction contain different types of TE elements (Figure 2a). The different types of TE elements are defined as two different TE classes, or different orders in a class or different super-family in an order. (ii) If the junctions result from an unknown sequence that separates two repeat sequences, these two repeat sequences must belong to two different types of TEs (Figure 2b). If they are the same type of TE, then, at least, one hit at the junction region must start from the beginning or the end of a repeat element. Matching to the beginning or end of a repeat element allows for accurate determination of TE insertion sites. (iii) If a potential junction has an unknown sequence at one side (Figure 2c), the hit at the junction site must start from the beginning or the end of the repeat sequence (4). If junctions are caused by an insertion of one TE into another TE of different type, the hit of inserted TE at the junction site must start from the beginning or end of a repeat element (Figure 2d).
Primer design strategy
In addition to RJM, four other popular repeat junction-based primer design strategies have been implemented in RJPrimers as well. In the RJM system, one PCR primer must span a repeat junction, making this primer junction specific and unique in the genome. The other primer can be picked from any sequence region or a different repeat match region (Figure 3a). In the RJJM system, two repeat junctions are required and each primer must span a junction site to increase primer specificity (10) (Figure 3b). RBIP and IRAP are initially used to develop markers from retrotransposons, specifically from LTR TEs, but they can be extended to use DNA transposons as well (8). Those latter two marker systems plus ISBP can be categorized into the same group. One primer is picked from a repeat element, but the primer does not span a junction site. However, the junction site must be included in PCR products. Therefore, the second primer can be picked from different repeat region (IRAP and ISBP) (Figure 3c and e), or unique gene region (RBIP) (Figure 3d). IRAP can use only the second type of TE junction structure (Figure 1b), while the third type of junction structures (Figure 2C) is suitable for RBIP. Both RJM and ISBP can use all types of repeat junction structures. The sole difference between RJM and ISBP is that one primer in RJM spans a junction site, but does not in ISBP.
Figure 3.
Schematic primer design strategies of different TE junction-based primers. (a–e) represent different types of repeat junction-based markers as indicated in the right side of each diagram.
Web server and command line based pipeline for junction identification and primer design
The web server of RJPrimers consists of a set of CGI-based programs written in Perl and a Java program ‘RJFinder’, which can run on Linux or Solaris operating system using an Apache HTTP server and Perl interpreter program. A user friendly interface similar to that of BatchPrimer3 (18) was applied to provide users with a sequence input mechanism, repeat database selection, and flexible parameter settings for junction identification and primer design. A total of 16 repeat databases, including RepBase, MIPS REdata, TREP, maize TEDB and 12 TIGR plant repeat databases, are available. More repeat databases can be easily embedded into the web application and the databases can be periodically updated if new repeat sequences are compiled into the databases. The Primer3 core program (19) was employed to pick the best pairs of standard PCR primers and an additional primer-picking algorithm in BatchPrimer3 (18) was included to select position-restricted primers like repeat junction flanking primers in RJM.
As in BatchPrimer3, DNA sequences in FASTA format can be taken as input by either a copy-and-paste mechanism into the sequence text box or by uploading a sequence file. RJPrimers automatically removes any unrecognized characters from the sequences. To balance the workload on the server, a limit of 200 short sequences (up to 1.5 kb) is set on the RJPrimers web server. The command line based pipeline can serve for long sequences and large amount of sequence reads.
RJPrimers generates primer design output: (i) a main HTML page containing the primer design summary of all input sequences, (ii) an HTML table page and a tab-delimited text file listing all designed primers and their properties and (iii) a detailed primer view page for each sequence with successfully designed primers. The primer list can be saved as a text file or an MS Excel file for further editing or primer ordering. All pages generated can be downloaded as a ‘zip’ compressed file. The RJPrimers web server is available at http://wheat.pw.usda.gov/demos/RJPrimers.
The command line-based pipeline provides the capability to process a large amount of sequence data without memory and network speed limitations. The user-defined repeat databases can be integrated into the RJPrimers pipeline if the databases are recompiled into the RJPrimers- required format (see online user’s manual). The parameters for repeat junction identification and primer design need to be set up in the pipeline program before execution. All output files are saved in a directory for each execution. The command line-based programs and user’s manual together with recompiled repeat databases can be downloaded from the RJPrimers website at http://wheat.pw.usda.gov/demos/RJPrimers.
MATERIALS AND METHODS
Genomic sequences
To assess the uniqueness and abundance of TE junction-based markers, genome sequences of rice and Sanger shotgun sequences of the ancestor of hexaploid wheat D genome, A. tauschii, were downloaded for in silico and wet lab PCR experiments. Sequences of the rice genome (Oryza sativa L.ssp. indica) were downloaded from http://rise.genomics.org.cn/rice/link/download.jsp. The available ∼5000 Sanger shotgun sequences of A. tauschii were downloaded from the NCBI GSS database (http://www.ncbi.nlm.nih.gov/). To examine effectiveness of Roche 454 reads in repeat junction marker development, genomic sequences of A. tauschii accession AS8/78 (∼0.3× genome equivalent) were generated using the Roche 454 sequencing platform.
In silico analysis of TE junction-based primers
An independent program, electronic PCR (20,21), was employed to examine the uniqueness of repeat junctions and RJM primers. First, genome sequences were randomly cut into 300–700 bp non-overlapping fragments to simulate short sequences. Repeat junction identification and primer design were performed using the RJPrimers pipeline program. Designed primers were used to search against the hashed database created from the whole genome sequence. The maximum PCR product length was set to 1 kb and no mismatch or gaps were allowed in primer alignments. The primer pair is counted as unique if only one in silico PCR product for a pair of TE junction-based primers was found in the search against the whole genome.
Wet lab evaluation of RJM primers
A random sample of 55 RJM primer pairs from 37 102 primer pairs designed from Roche 454 reads of A. tauschii and 16 primer pairs from 244 primer pairs designed from A. tauschii Sanger shotgun sequences were chosen for verification by PCR. In addition, 25 primer pairs were manually designed from the same set of sequences through manual BLAST searches and primer picking to compare effectiveness of the automated and manual approaches. The default primer-designing parameters used in RJPrimers were as follows: primer length of 19–22 bases with the optimum 20 bases, Tm of 55–63°C with the optimum 58°C, GC content of 40–60%, a 300 nt optimum product size with a range from 150 to 700 nt. Several other restrictions for primer specificity were also superimposed, including the maximum primer self complementarity (whole primer) of 5.0, the maximum 3′ self complementarity of 2.0. PCR was performed in a total volume of 20 µl with 5 mmol/l of each dNTP, 5 mmol/l of each primer, 0.2 units GoTaq polymerase (Promega, Madison, Wisconsin) with 5× buffer, and 100 ng template DNA. The temperature regime consisted of a 4 min initial denaturation step at 94°C, followed by 35 cycles of 94°C for 20 s, 57°C for 20 s and 72°C for 60 s, and a final extension at 72°C for 5 min. PCR products were then separated on 2.5% Metaphor agarose 1× TAE gels (Cambrex Bio-Science Inc., Rockland, Maine).
RESULTS AND DISCUSSION
Abundance and uniqueness of repeat junction markers
Genome-wide repeat junctions were identified by RJPrimers in rice (Table 1). A total of 59 182 repeat junctions were identified in the 401 Mb sequences. On average, 0.148 junctions per kilobase (Kb) were detected. Since the rice sequence was cut into 300–700 bp fragments to simulate short sequences, some junctions which were close to the ends of fragments were likely excluded by RJPrimers. To assess the effects of this factor, the chromosomes in full length were used. A total of 0.328 junctions per Kb were obtained, two times more junctions than from short sequences. In A. tauschii, 0.183 junctions per Kb for random shotguns Sanger sequences and 0.113 junctions per Kb for the Roche 454 reads were identified. Aegilops tauschii has a higher repeat percentage (∼90%) (22) than rice (∼35%), thus more junctions than in rice are to be expected. A lower repeat junction rate in Roche 454 reads than Sanger shotgun sequences is due to the shorter read length (366 bp on average) in Roche 454 reads as compared to Sanger shotgun sequences (730 bp). However, with the much higher sequencing throughput by the Roche 454, we can expect that the 454-based sequencing technology will be more useful in developing RJM for complex genomes with limited genomic sequence information.
Table 1.
Abundance and uniqueness of repeat junctions and RJM primers
| Source | Sequence size (Mb) | No. of repeat junctions identified | No. of junctions per kb | No. of RJM markers designed | Unique RJM markers (%) | Unique RJM markers per kb |
|---|---|---|---|---|---|---|
| Oryza sativa L. ssp. indica genome | 401 | 59 182 | 0.148 | 16 280 | 90.8 | 0.041 |
| Aegilops tauschii shotgun sequences | 3.7 | 674 | 0.183 | 244 | 92.3a | 0.169a |
| Aegilops tauschii Roche 454 reads (0.3× genome equivalent) | 1093.6 | 123 683 | 0.113 | 37 102 | 90.7a | 0.102a |
aEstimated from wet lab PCR amplification of a random sample (see details in Table 2).
TE junctions can be easily converted into PCR-based markers (11). A total of 16 280 primer pairs (28%) were designed from 59 182 rice repeat junctions and 90.8% of RJM primers or 0.041 primers per Kb were unique, as indicated by single in silico PCR products (Table 1 and Figure 4). The in silico PCR could help users evaluate the uniqueness of RJ markers generated in the targeted genome provide the genome sequence information is available. Taken together, we can expect to produce at least four unique sets of RJM primer pairs per 100 Kb in genomes with repeat content similar to that of rice. Not all repeat junctions can be used for RJM primer design due to certain primer design restrictions. Approximately 30% of junctions identified from rice and A. tauschii were usable since high stringent cutoff values were imposed on PCR primer design parameters as default in RJPrimers in order to increase the success rate of PCR amplification.
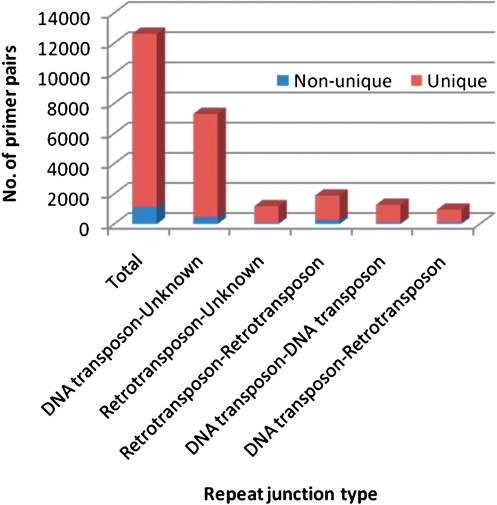
Figure 4.
Distribution of unique and non-unique primers designed from rice genome (Oryza sativa L. ssp indica) in different types of repeat junctions.
For the repeat junctions identified in rice, most junctions were grouped into the ‘DNA transposon—unknown’ category (Figure 4). This is in agreement with the notion that DNA transposons greatly outnumber retrotransposons in rice although retrotransposons account for larger portion of nucleotides than DNA transposon (1). The ‘unknown’ sequence regions were most likely unique intergenic or gene regions. By examining unique primers in different repeat categories, DNA transposon- and retrotransposon-unknown repeat junctions generated more unique RJM primers (93%) than the other three categories (∼87%) in rice. Repeat junctions between the same types of TEs contributed to 25% of all non-unique primers, but to only 15% of all unique primers. Therefore, a manual review of primers designed from retrotransposon–retrotransposon and DNA transposon–DNA transposon junctions is recommended to reduce the false positive junctions.
Because A. tauschii genome has not been sequenced, a random sample of 71 primer pairs from Roche 454 reads and Sanger shotgun sequences were used to perform wet lab PCR amplification for evaluating uniqueness of RJM primers designed by RJPrimers. This also allowed us to examine if the high-throughput sequencing by Roche 454 can be effectively used for developing repeat junction markers in complex genomes. Of 71 primer pairs, 67 primer pairs (94.6%) produced successful amplification (Table 2, Figure 5) and only four primer pairs failed to amplify A. tauschii DNA. Among 67 amplified primer pairs, 61 primer pairs (91%) generated clear single bands, indicating that these primers and repeat junctions are most likely unique because most of those repeat junction markers with single bands were unambiguously mapped to individual wheat chromosomes and delineated bins of the chromosomes (5). There were six primer pairs which amplified two or three bands. The rate of unique primers in A. tauschii (90.7 and 92.3%) was similar to the rate in rice (90.8%) (Table 1), and it is also consistent with the rate estimated from 300 RJM primers designed semi-automatically from BAC end and shotgun Sanger sequences of A. tauschii (90%) (5). The RJM primers from Roche 454 reads had a similar rate of unique primers with that from shotgun Sanger sequences (Tables 1 and 2).
Table 2.
Wet lab PCR amplification of RJM primers designed by RJPrimers from Roche 454 reads and Sanger shotgun sequences of A. tauschii
| No. of primers designed | No. of primers with amplification |
No. of primers failed to amply | ||
|---|---|---|---|---|
| Single product | Multiple products | |||
| Sequence sources | ||||
| Roche 454 reads | 55 | 49 | 5 | 1 |
| Shotgun sequences | 16 | 12 | 1 | 3 |
| Total | 71 | 61 | 6 | 4 |
| Repeat junction categories | ||||
| Retrotransposon–retrotransposon | 24 | 22 | 2 | 1 |
| Retrotransposon–DNA transposon | 19 | 16 | 2 | 1 |
| Retrotranspson–unknown | 19 | 17 | 1 | 1 |
| DNA transposon–DNA transposon | 4 | 3 | 0 | 1 |
| DNA transposon unknown | 5 | 4 | 1 | 0 |
| Total | 71 | 62 | 6 | 4 |
Figure 5.
PCR amplification using RJM primers designed by RJPrimers and A. tauschii genomic DNA. The DNA sequences used for primer design were Roche 454 reads from A. tauschii genomic DNA. A single PCR product (single band) indicates that the primer pair amplifies unique marker. The image shows the amplification products of 19 primer pairs.
Twenty-five repeat junction primer pairs were manually designed in order to compare the automatic RJPrimers approach with manual one. Of 25 primer pairs, 19 primer pairs (76%) produced successful amplification, and six primers (24%) failed to amply, suggesting a higher failure rate than the automatic approach (four primer pairs, 5.6%). In those primers which failed, one primer pair failed in both manual and automatic approaches, two primer pairs failed in manual design but worked in the automated approach. The other three primer pairs were designed from different sequences. Usually manual identification of repeat junction using BLASTN searches may yield more repeat junctions because RJPrimers uses stringent criteria to filter out false positive repeat junctions. Therefore, some true repeat junctions are also excluded. However, manual primer design is inefficient and no consistent primer properties are applied to primer design. The automatic RJM primer design by RJPrimers appears therefore to be superior to the manual approach in efficiency and success rate of PCR amplification.
Aegilops tauschii repeat junction categories differed from those in rice (Table 2). Most of the repeat junctions in A. tauschii involved retrotransposons rather than DNA transposons, as was the case in rice. This is because 75% of repeat reads are retrotransposons based on repeat annotation in the ∼0.3× A. tauschii genome coverage produced by Roche 454 reads. Although the two grass species differ in frequencies of repeat junction categories, they both have a similarly high level of unique repeat junctions and RJM primers.
Comparison of different repeat junction-based primer design strategies
Five different TE junction-based primer design strategies were implemented in RJPrimers. To compare those five strategies, chromosome 1 of the rice genome (Oryza sativa L. ssp indica) was used for in silico analysis. A total of 4234 junctions were found. From those junction regions, five different types of primers were designed (Table 3). Only one primer pair was picked per repeat junction. From the 4234 predicted repeat junctions, RJM and ISBP generated high numbers of primer pairs, 1897 (45%) and 1837 (43%), respectively, using the default settings of RJPrimers, while RJJM and IRAP generated the lowest numbers of primers (28 and 30, respectively). The large difference in primer numbers generated from the same number of junctions is due to the primer design strategy itself and the distribution of the repeat junction types in the chromosome. RJM and ISBP are therefore the most flexible design strategies because they can make full use of all possible types of repeat junctions (Figure 2). RJJM and IRAP must use two junctions to design primers but most of short sequences contain only one junction and fail to allow the design for those types of markers. Electronic PCR analysis showed that all types of primers have a similar, high percentage of unique primers (>90%). RJJM and IRAP had the highest percentages of unique primers (93%), likely due to the use of two unique junctions (Figure 3). Previous studies (4,5) showed that RJM and ISBP markers are genome-specific, unique, allelic, and useful in genetic, physical and radiation mapping in the hexaploid wheat. In silico and wet lab PCR tests in this study consistently confirmed that RJM and ISBP primers are the most abundant and unique both in rice and wheat. Thus, RJM and ISBP are two high-throughput and unique marker systems.
Table 3.
Comparison of different types of TE junction based primers using e-PCR
| Primer type | No. of repeat junctionsa | No. of designed primer pairs | Unique primer pairs (%) | Unique primer pairs per Kb |
|---|---|---|---|---|
| RJM | 4234 | 1897 | 91.8 | 0.038 |
| RJJM | 4234 | 28 | 92.9 | 0.001 |
| ISBP | 4234 | 1837 | 91.6 | 0.037 |
| RBIP | 4234 | 443 | 90.3 | 0.009 |
| IRAP | 4234 | 30 | 93.0 | 0.001 |
aChromosome 1 sequence of Oryza sativa L. ssp indica was used for repeat junction identification and primer design.
Performance of RJPrimers
Two versions of RJPrimers are provided for web access and command line processing. RJPrimers includes three contiguous operational steps, a BLASTN search against a repeat database, repeat junction identification and primer design. For web-based RJPrimers, one additional step is required—loading sequence data through a network to the web server. Client Internet speed will affect sequence loading. Therefore, the performance of RJPrimers depends on the number of sequences and their sizes, the number of repeat junctions in the sequences, the size of the repeat database selected, and the speed of the computer, and the Internet speed for the web version. As a simple benchmark, a total of 5055 A. tauschii shotgun sequences and the TREP database were used for RJM primer design. It took a total of 4.08 min for finishing the entire pipeline on a desktop computer (Asus P6T, Intel core i7 920, 12 GB of RAM, and a Ubuntu Linux 9.04 64 bit operating system).
FUNDING
This work is supported in part by US National Science Foundation (grant numbers IOS 0701916 and IOS 0822100) and by US Department of Agriculture, Agriculture Research Service CRIS projects 5325-21000-014. Funding for open access charge: US Department of Agriculture, Agriculture Research Service.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We thank Roger Thilmony and Xiaohua He for the critical reading of the manuscript. We are very grateful to Farhad Ghavami, Ajay Kumar, Vijay Tiwari, Jiajie Wu and the anonymous reviewers for testing the server and offering valuable comments
REFERENCES
- 1.Turcotte K., Srinivasan S., Bureau T. Survey of transposable elements from rice genomic sequences. Plant J. 2001;25:169–179. doi: 10.1046/j.1365-313x.2001.00945.x. [DOI] [PubMed] [Google Scholar]
- 2.Dvorak J. Triticeae genome structure and evolution. In: Feuillet C., Muehlbauer G.J., editors. Genetics and Genomics of the Triticeae. New York, USA: Springer; 2009. pp. 685–711. [Google Scholar]
- 3.Wicker T., Sabot F., Hua-Van A., Bennetzen J.L., Capy P., Chalhoub B., Flavell A., Leroy P., Morgante M., Panaud O., et al. A unified classification system for eukaryotic transposable elements. Nat. Rev. Genet. 2007;8:973–982. doi: 10.1038/nrg2165. [DOI] [PubMed] [Google Scholar]
- 4.Paux E., Roger D., Badaeva E., Gay G., Bernard M., Sourdille P., Feuillet C. Characterizing the composition and evolution of homoeologous genomes in hexaploid wheat through BAC-end sequencing on chromosome 3B. Plant J. 2006;48:463–474. doi: 10.1111/j.1365-313X.2006.02891.x. [DOI] [PubMed] [Google Scholar]
- 5.Wanjugi H., Coleman-Derr D., Huo N., Kianian S.F., Luo M.C., Wu J., Anderson O., Gu Y.Q. Rapid development of PCR-based genome-specific repetitive DNA junction markers in wheat. Genome. 2009;52:576–587. doi: 10.1139/g09-033. [DOI] [PubMed] [Google Scholar]
- 6.Paux E., Faure S., Choulet F., Roger D., Gauthier V., Martinant J.P., Sourdille P., Balfourier F., Le Paslier M.C., Chauveau A., et al. Insertion site-based polymorphism markers open new perspectives for genome saturation and marker-assisted selection in wheat. Plant Biotechnol. J. 2010;8:196–210. doi: 10.1111/j.1467-7652.2009.00477.x. [DOI] [PubMed] [Google Scholar]
- 7.Waugh R., McLean K., Flavell A.J., Pearce S.R., Kumar A., Thomas B.B., Powell W. Genetic distribution of Bare-1-like retrotransposable elements in the barley genome revealed by sequence-specific amplification polymorphisms (S-SAP) Mol. Gen. Genet. 1997;253:687–694. doi: 10.1007/s004380050372. [DOI] [PubMed] [Google Scholar]
- 8.Kalendar R., Schulman A.H. IRAP and REMAP for retrotransposon-based genotyping and fingerprinting. Nat. Protoc. 2006;1:2478–2484. doi: 10.1038/nprot.2006.377. [DOI] [PubMed] [Google Scholar]
- 9.Flavell A.J., Knox M.R., Pearce S.R., Ellis T.H. Retrotransposon-based insertion polymorphisms (RBIP) for high throughput marker analysis. Plant J. 1998;16:643–650. doi: 10.1046/j.1365-313x.1998.00334.x. [DOI] [PubMed] [Google Scholar]
- 10.Luce A.C., Sharma A., Mollere O.S., Wolfgruber T.K., Nagaki K., Jiang J., Presting G.G., Dawe R.K. Precise centromere mapping using a combination of repeat junction markers and chromatin immunoprecipitation-polymerase chain reaction. Genetics. 2006;174:1057–1061. doi: 10.1534/genetics.106.060467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Devos K.M., Ma J., Pontaroli A.C., Pratt L.H., Bennetzen J.L. Analysis and mapping of randomly chosen bacterial artificial chromosome clones from hexaploid bread wheat. Proc. Natl Acad. Sci. USA. 2005;102:19243–19248. doi: 10.1073/pnas.0509473102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McCarthy E.M., McDonald J.F. LTR_STRUC: a novel search and identification program for LTR retrotransposons. Bioinformatics. 2003;19:362–367. doi: 10.1093/bioinformatics/btf878. [DOI] [PubMed] [Google Scholar]
- 13.Xu Z., Wang H. LTR_FINDER: an efficient tool for the prediction of full-length LTR retrotransposons. Nucleic Acids Res. 2007;35:W265–W268. doi: 10.1093/nar/gkm286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bao Z., Eddy S.R. Automated de novo identification of repeat sequence families in sequenced genomes. Genome Res. 2002;12:1269–1276. doi: 10.1101/gr.88502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wolfgruber T.K., Presting G.G. JunctionViewer: customizable annotation software for repeat-rich genomic regions. BMC Bioinformatics. 2010;11:23. doi: 10.1186/1471-2105-11-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Spannagl M., Haberer G., Ernst R., Schoof H., Mayer K.F. MIPS plant genome information resources. Methods Mol. Biol. 2007;406:137–159. doi: 10.1007/978-1-59745-535-0_6. [DOI] [PubMed] [Google Scholar]
- 17.Ouyang S., Buell C.R. The TIGR Plant Repeat Databases: a collective resource for the identification of repetitive sequences in plants. Nucleic Acids Res. 2004;32:D360–D363. doi: 10.1093/nar/gkh099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.You F.M., Huo N., Gu Y.Q., Luo M.C., Ma Y., Hane D., Lazo G.R., Dvorak J., Anderson O.D. BatchPrimer3: a high throughput web application for PCR and sequencing primer design. BMC Bioinformatics. 2008;9:253. doi: 10.1186/1471-2105-9-253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rozen S., Skaletsky H.J. Primer3 on the WWW for general users and for biologist programmers. In: Krawet S., Misener S., editors. Bioinformatics Methods and Protocols: Methods in Molecular Biology. Totowa, NJ, USA: Humana Press; 2000. pp. 365–386. [DOI] [PubMed] [Google Scholar]
- 20.Schuler G.D. Sequence mapping by electronic PCR. Genome Res. 1997;7:541–550. doi: 10.1101/gr.7.5.541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rotmistrovsky K., Jang W., Schuler G.D. A web server for performing electronic PCR. Nucleic Acids Res. 2004;32:W108–W112. doi: 10.1093/nar/gkh450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li W., Zhang P., Fellers J.P., Friebe B., Gill B.S. Sequence composition, organization, and evolution of the core Triticeae genome. Plant J. 2004;40:500–511. doi: 10.1111/j.1365-313X.2004.02228.x. [DOI] [PubMed] [Google Scholar]