Abstract
One of the greatest challenges in bioinformatics is to shed light on the relationship between genomic and chemical significances of metabolic pathways. Here, we demonstrate two types of chemical structure search servers: SIMCOMP (http://www.genome.jp/tools/simcomp/) for the chemical similarity search and SUBCOMP (http://www.genome.jp/tools/subcomp/) for the chemical substructure search, where both servers provide links to the KEGG PATHWAY and BRITE databases. The SIMCOMP is a graph-based method for searching the maximal common subgraph isomorphism by finding the maximal cliques in the association graph. In contrast, the SUBCOMP is an extended method for solving the subgraph isomorphism problem. The obtained links to PATHWAY or BRITE databases can be used to interpret as the biological meanings of chemical structures from the viewpoint of the various biological functions including metabolic networks.
INTRODUCTION
To understand the complex system of life it is required to investigate the characteristics of biomolecular network consisting of not only proteins or nucleic acids but also small chemical compounds (1,2). In spite of a number of sequence comparison servers, there are few servers to compare chemical structures for metabolic pathway analyses. In addition, almost all servers for chemical structure comparisons commonly use the bit-string method (3), and there had been no server based on the graph comparison method, which is considered more accurate. In this context, we have developed two types of chemical structure search servers as parts of the GenomeNet computation services, aiming at the better comprehension of the relationship between genomic and chemical implications of metabolic pathways. One is the SIMCOMP (4,5) server for the chemical similarity search and the other is the SUBCOMP for the chemical substructure search, where both servers provide links to the KEGG PATHWAY and BRITE databases (6). SIMCOMP (SIMilar COMPound) has originally been developed as a graph-based method for comparing chemical structures, which searches for the maximal cliques in the association graph as the maximum common induced subgraph (MCIS). However, the current version of SIMCOMP can also compute the maximum common edge subgraph (MCES), which is faster because of the small number of nodes in an association graph. Moreover, we have now added further computation features to SIMCOMP, including chirality check and PATHWAY/BRITE mapping. In contrast, SUBCOMP (SUBstructure matching of COMPounds) is an extended method based on the bit-vector representation for searching chemical substructures, which is often used as a rapid alternative to more time-consuming (but more accurate) SIMCOMP.
The notable features of the SIMCOMP and SUBCOMP servers are as follows: (i) After obtaining the list of similar compounds, users can map the selected entries onto the KEGG PATHWAY or KEGG BRITE databases. This feature may help us to investigate biological roles of those chemical compounds as well as a query compound. (ii) Both SIMCOMP and SUBCOMP can discriminate the isomeric structures, including the R−/S− chirality found at asymmetrical carbons and the cis–trans isomerism around the carbon–carbon double bonds. (iii) Various matching conditions are now available in the SUBCOMP computation. ‘Charge’ and ‘Valence’ options will distinguish ionized atoms from normal atoms and the valence of each atom, in other words, the oxidation state of each atom, respectively. ‘Coordinate bond’ option can be used to consider the coordinate bond formed between anion–cation single bond. (iv) The superstructure search is also available for searching chemical compounds that are included in the query structure, in the SUBCOMP.
With these characteristic features, both structure search servers can provide the way of the biochemical analyses on the metabolic networks for chemical compounds including bioactive natural products and drugs. The results of mapping onto PATHWAY or BRITE databases may indicate which biological functions are involved in the selected chemicals.
IMPLEMENTATION AND USAGE
Common features
Query structure
A query compound structure can be one of the following four formats: (i) an MDL mol file (7) saved on the local machine, (ii) direct input of MDL mol format into the textarea, (iii) SMILES representation (8) and (iv) an entry identifier, C or D number, of KEGG COMPOUND or DRUG databases (9). In case of the query structure specified by C or D number or in the MDL mol format, the user can preview the query structure on screen before an actual computation, otherwise no preview functionality is provided since there is no information about x–y coordinates. The SIMCOMP server converts the input chemical structure into the KEGG Chemical Function (KCF) format internally, which is one of the most prominent heuristics. In the KCF format, all atoms are represented as the KEGG Atom Types, which are based on the concept of functional groups in organic chemistry, and 68 atom types (vertex types) have been defined for carbon, nitrogen, oxygen, sulfur, phosphorus and other atomic species with different environments (4,5).
The user can easily change the computation algorithm between SIMCOMP and SUBCOMP by clicking the tabs on the top of the query form.
Target databases
The current target database is one of the following four databases: (i) KEGG COMPOUND for chemical compounds in metabolic pathways and other biologically related compounds, (ii) KEGG DRUG for the approved drug structures, (iii) KNApSAcK for chemical compounds mainly from secondary metabolisms (10) and (iv) KEGG REACTION for chemical compounds in enzyme-catalyzed and spontaneous reactions in biological systems (9).
Output of computation
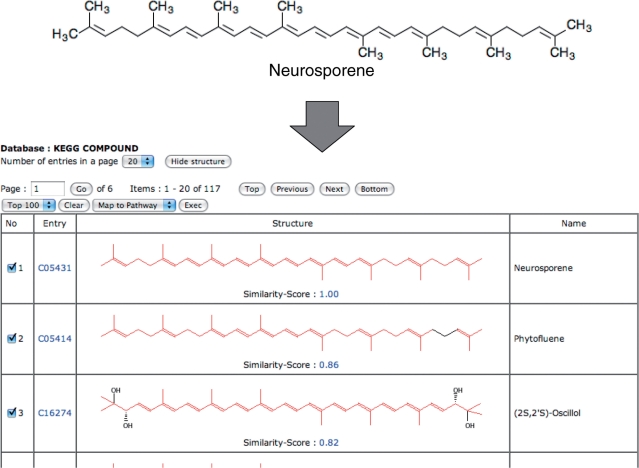
After calculations, both servers will output the list of database entries similar to (SIMCOMP) or having the same substructures with (SUBCOMP) the query structure. The information of atom alignments to the query structure is provided to each database entry. The similarity between the query and the database entry can be estimated by the number of matched atoms against the total number of matched and unmatched atoms from the atom alignment. Consequently, the definition of similarity scores between two chemical graphs can be formulated in the similar manner of Tanimoto coefficient between two bit-represented vectors, which has been used as one of the most famous and succeeded proximities in the chemical structure search systems (3,11). The database entries are listed in descending order of similarity scores by default (Figure 1).
Figure 1.
The examples of an input query and computation results. The upper section is the input query ‘C05431’ (neurosporene), which is a precursor of lycopene, and the lower section is the computation results by SIMCOMP for the COMPOUND database with the ‘global search’ option.
Chiral check
The chirality difference between query compound and database entry is tested when the chirality information is properly given. Here, the R−/S− chirality of asymmetric carbons can be designated as the up or down arrows on the 2D graph, and the cis−/trans− chirality around the carbon–carbon double bond should be described with correct x–y coordinates on 2D plane. When the chirality difference is detected, the resulting similarity scores will be recalculated by penalizing each difference by 0.1 atom match to distinguish the isomers.
Mapping to KEGG PATHWAY and BRITE
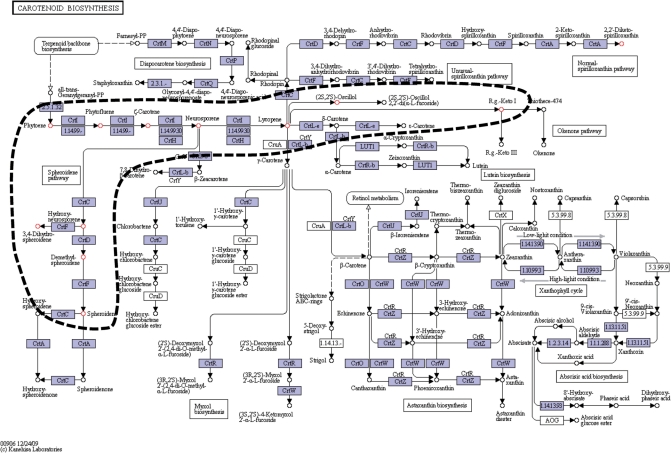
After obtaining the search result against KEGG databases (COMPOUND, DRUG and REACTION), the user can further map the selected entries onto the KEGG PATHWAY or KEGG BRITE databases by choosing ‘Map to Pathway’ or ‘Map to BRITE’ from the ‘Select operation’ menu, respectively. When mapping onto PATHWAY or BRITE database, the related pathway maps or BRITE hierarchies that contain at least one of the selected entries are listed with links to the actual pathway maps or BRITE entries. The selected chemical compounds are emphasized with color and Figure 2 shows an example where the compounds are indicated by red circles.
Figure 2.
The example of the further mapping onto metabolic pathways. An area, where the compounds similar with C05431 (neurosporene) have been frequently found, can be easily detected by the red colored compounds (indicated by the dashed line in this figure). This means that a neurosporene and its derivatives are likely to be located around the gateway of the ‘Carotenoid biosynthesis’ pathway (map00906). Actually, they are important intermediates for generating carotenoids.
SIMCOMP-specific features
The computation of SIMCOMP is based on the efficient algorithm to find the maximum common substructures between two given chemical structures represented as 2D graphs consisting of atoms as vertices and covalent bonds as edges. The algorithm is implemented by the program finding the maximal cliques in the association graph of two graphs as the MCIS or as the MCES. SIMCOMP adopts several heuristics to decrease the computation difficulty of clique finding as well as to increase the chance of biologically meaningful matches by using KEGG Atom Types. The user can choose these heuristics from the advanced options.
Advanced options
The user can select the method to make the association graph, that is, the atom-based approach (MCIS) or the bond-based approach (MCES). MCES checks all possibilities of matching four atoms connected by each bond. This means that the MCES is stricter than MCIS, resulting in the smaller association graph and the faster clique finding. In usual cases, MCES is about tenth faster than MCIS. However, MCES does not produce a result with only one atom match.
The user can also select the post-processing treatment to make a full alignment (maximal common subgraphs) from several simply connected common subgraphs (SCCSs). The option ‘for the largest SCCS’ only keeps the largest SCCS and extends it. Another option ‘for all SCCSs’ keeps all SCCSs and tries to connect them.
The node conditions of making the association graph and of extending the SCCSs can be controlled by selecting one of three levels: (i) the atom species like ‘C’, (ii) the class of KEGG Atom Types, like ‘C1′ or (iii) the whole notation of KEGG Atom Types, like ‘C1a’.
To simplify the setting of the above various search conditions, we provide the two major search settings as Global and Local search for capturing the global tendency of chemical similarity and the local structural matching, respectively.
SUBCOMP-specific features
The SUBCOMP is a novel algorithm for solving the subgraph isomorphism problem by extending Ullmann’s algorithm (12), which tries to assign every possible node of the query graph to each node of the target graph by recursively generating a match matrix. A technique called refinement is adopted in every recursion step in order to reduce the number of matching candidates. In the refinement step, the validity of match matrices can be checked using bitwise operations, in which 64 atoms can be checked simultaneously with 64-bit microprocessors. This means the refinement technique is very fast and appropriate for usual microprocessors. In our implementation of SUBCOMP, we have further optimized the refinement step in Ullmann’s algorithm by compiling and storing adjacency matrices of target molecules as the bit-vector database. This greatly decreased the computational time of the database search. In comparison with other algorithms (13–16) such as Vento-Foggia (VF) algorithm (17), the bitwise operations of the SUBCOMP or the Ullmann’s algorithm may work efficiently for searching the graphs having relatively small number of nodes (such as chemical compound graphs). Although the VF algorithm will be still more efficient for very large graphs, we have chosen the Ullmann’s algorithm based method in the current implementation, because most compounds are relatively small and the implementation is very easy.
Consequently, the computation time is about a few seconds on our server for the queries in the size of actual metabolites found in KEGG. Hence, SUBCOMP can be a faster alternative to more time-consuming (but more accurate) SIMCOMP for searching similar chemical structures. The limitation of the SUBCOMP is that the algorithm only checks whether the whole atoms and bonds of the query graph are included within the target graph or not, in contrast to finding the maximal common subgraph between two graphs. This indicates that the similarity scores of computation results do not mean the similarities of whole chemical structures but the partial contribution of substructures. The current version of SUBCOMP can identify the database entries containing substructures that match the entire query compound and/or the database compounds that match any substructure of the query compound.
Advanced options
When encoding the bit-vector representation for each atom, the SUBCOMP can incorporate the following three atom conditions: ionization, valence and coordinate bond. For the ionization of atoms, sometimes the oxygen of a hydroxyl group (‘OH’) is described as the anion like ‘O−’ for the scientific strictness of molecular representations. In this case, the usual algorithm will be failed to compare these two oxygens, because of the existence of a hydrogen atom or a related bond. Moreover, the user may want to distinguish the valence or the oxidation state of each atom. For instance, in the case of sulfur atoms, the different valences in H2SO4 (+6) and H2S (−2) should be distinguished from the viewpoint of searching more biochemically meaningful substructures. The coordinate bond will also be considered when there is a single covalent bond between anion and cation atoms. In this case, such a bond can be aligned with a normal double bond.
Since the size of the bits-vectors used in the advanced options is the same as those for the default parameters, the computation speed does not change.
CONCLUSIONS
We have developed two types of chemical structure search servers; one is the similarity search server and another is the subgraph search, which is much faster than the former one. Both servers can provide the opportunity to investigate the detailed chemical features, including chirality and other physicochemical properties. A current limitation is the handling of R-group representation. Because many databases do not distinguish R-group representations for aromatic and aliphatic groups, SIMCOMP and SUBCOMP cannot distinguish them. The different scoring system for such R groups from other atoms would improve the system.
SIMCOMP and SUBCOMP should have an advantage in the further comprehension of the biological functions of chemical compounds, which may be inferred from two types of mapping. The mapping results to metabolic networks indicate the biological significance of chemical structures as a part of successive reaction pathways, and the results of mapping to BRITE hierarchies may designate the classes of biological roles in each functional hierarchy. This may be helpful to grasp the biological meanings of the set of similar chemical compounds in the life system from the genomic viewpoint, because we can easily correlate the chemical compounds on the pathway map and related enzyme genes on the same pathway using the mapping result. Further implementation such as the comprehensive similarity search and pathway mapping using metabolome data as queries may be one of the most significant goals that should be achieved in the future.
Another possible extension is the matching problem between the ligand and the protein structure. This problem has been well described as the ligand docking for many years, where the structural complementarities between the ligand structure and the protein surface asperity are considered using the actual 3D coordinates. Our matching method does not require any 3D information on every compound, but we need only the topology of compound structures, which might be a big hurdle for the purpose of the docking simulation. However, when we can extract the information on the preferable interaction between each ligand atom and each protein atom from the 3D complex structures, we may give appropriate matching scores to each atom–atom matching between the ligand and the protein and then obtain the plausible docking configurations by the 2D graph matching.
AVAILABILITY
The current version of both servers has been available since 1 April 2009. The program package including the stand-alone versions of SIMCOMP and SUBCOMP as well as other related programs will be also available soon at the GenomeNet (http://www.genome.jp/).
FUNDING
Grants from the Ministry of Education, Culture, Sports, Science and Technology of Japan; Japan Science and Technology Agency. The computational resources were provided by the Bioinformatics Center, Institute for Chemical Research, Kyoto University. Funding for open access charge: Japan Science and Technology Agency.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We thank Tomohiro Ohya and Yusuke Sugihara for helpful discussions on the graph isomorphism problem of chemical compounds.
REFERENCES
- 1.Kanehisa M. Prediction of higher order functional networks from genomic data. Pharmacogenomics. 2001;2:373–385. doi: 10.1517/14622416.2.4.373. [DOI] [PubMed] [Google Scholar]
- 2.Oprea TI, Tropsha A, Faulon JL, Rintoul MD. Systems chemical biology. Nat. Chem. Biol. 2007;8:447–450. doi: 10.1038/nchembio0807-447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang Y, Xiao J, Suzek TO, Zhang J, Wang J, Bryant SH. PubChem: a public information system for analyzing bioactivities of small molecules. Nucleic Acids Res. 2009;37:W623–W633. doi: 10.1093/nar/gkp456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hattori M, Okuno Y, Goto S, Kanehisa M. Development of a chemical structure comparison method for integrated analysis of chemical and genomic information in the metabolic pathways. J. Am. Chem. Soc. 2003;125:11853–11865. doi: 10.1021/ja036030u. [DOI] [PubMed] [Google Scholar]
- 5.Hattori M, Okuno Y, Goto S, Kanehisa M. Heuristics for chemical compound matching. Genome Inform. 2003;14:144–153. [PubMed] [Google Scholar]
- 6.Kanehisa M, Araki M, Goto S, Hattori M, Hirakawa M, Itoh M, Katayama T, Kawashima S, Okuda S, Tokimatsu T, et al. KEGG for linking genomes to life and the environment. Nucleic Acids Res. 2008;36:D480–D484. doi: 10.1093/nar/gkm882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dalby A, Nourse JG, Hounshell WD, Gushurst A.KI, Grier DL, Leland BA, Laufer J. Description of several chemical structure file formats used by computer programs developed at Molecular Design Limited. J. Chem. Inf. Comput. Sci. 1992;32:244–255. [Google Scholar]
- 8.Weininger D. SMILES, a chemical language and information system. 1. Introduction to methodology and encoding rules. J. Chem. Inf. Comput. Sci. 1988;28:31–36. [Google Scholar]
- 9.Goto S, Okuno Y, Hattori M, Nishioka T, Kanehisa M. LIGAND: database of chemical compounds and reactions in biological pathways. Nucleic Acids Res. 2002;30:402–404. doi: 10.1093/nar/30.1.402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shinbo Y, Nakamura Y, Altaf-Ul-Amin M, Asahi H, Kurokawa K, Arita M, Saito K, Ohta D, Shibata D, Kanaya S. KNApSAcK: A comprehensive species-metabolite relationship database. Biotechnol. Agric. For. 2006;57:165–181. [Google Scholar]
- 11.Willett P, Barnard J, Downs GM. Chemical similarity searching. J. Chem. Inf. Comput. Sci. 1998;38:983–996. [Google Scholar]
- 12.Ullmann JR. An algorithm for subgraph isomorphism. J. ACM. 1976;23:31–42. [Google Scholar]
- 13.Golovin A, Henrick K. Chemical substructure search in SQL. J. Chem. Inf. Model. 2009;49:22–27. doi: 10.1021/ci8003013. [DOI] [PubMed] [Google Scholar]
- 14.Chen L, Nourse JG, Christie BD, Leland BA, Grier DL. Over 20 years of reaction access systems from MDL: a novel reaction substructure search algorithm. J. Chem. Inf. Comput. Sci. 2002;42:1296–1310. doi: 10.1021/ci020023s. [DOI] [PubMed] [Google Scholar]
- 15.Berks AH. Current state of the art of Markush topological search systems. In: Gasteiger J, editor. Handbook of Chemoinformatics: from Data to Knowledge. Vol. 2. Weinheim: Wiley-VCH; 2003. pp. 885–903. [Google Scholar]
- 16.Liliana F, Valiente G. Validation of metabolic pathway databases based on chemical substructure search. Biomol. Eng. 2007;24:327–335. doi: 10.1016/j.bioeng.2007.02.008. [DOI] [PubMed] [Google Scholar]
- 17.Cordella LP, Foggia P, Sansone C, Vento M. Proceedings of the 13th ICPR. Vol. III. Washington, DC: IEEE Computer Society Press; 1996. An efficient algorithm for the inexact matching of ARG using a contextual transformational model; pp. 180–184. [Google Scholar]