Abstract
The energy distribution along the protein–protein interface is not homogenous; certain residues contribute more to the binding free energy, called ‘hot spots’. Here, we present a web server, HotPoint, which predicts hot spots in protein interfaces using an empirical model. The empirical model incorporates a few simple rules consisting of occlusion from solvent and total knowledge-based pair potentials of residues. The prediction model is computationally efficient and achieves high accuracy of 70%. The input to the HotPoint server is a protein complex and two chain identifiers that form an interface. The server provides the hot spot prediction results, a table of residue properties and an interactive 3D visualization of the complex with hot spots highlighted. Results are also downloadable as text files. This web server can be used for analysis of any protein–protein interface which can be utilized by researchers working on binding sites characterization and rational design of small molecules for protein interactions. HotPoint is accessible at http://prism.ccbb.ku.edu.tr/hotpoint.
INTRODUCTION
Most molecular and cellular processes are controlled by protein–protein interactions. Proteins interact through interfaces. The energy distribution along the interface region is not homogenous; certain residues contribute more to the binding free energy, called ‘hot spots’ (1–3). Hot spots form tightly packed regions in protein interfaces (4). Presence of hot spots is important as a target to disrupt malfunctioning association of proteins by therapeutic molecules and for rational design of highly specific protein complexes (5,6). Experimentally, a hot spot can be found by evaluating the change in binding free energy upon mutating it to an alanine. For a limited number of protein complexes, alanine mutations are available and this information is deposited in databases (7,8). Highly efficient computational methods emerged to identify hot spots under the occurrence of limitations in experimental information. Although there is not a strict rule to identify hot spots, combination of several physical and chemical features of residues gives successful results. Several groups developed energy based methods (9–12), learning based methods (13–19) and molecular dynamics based methods (20–22) to predict hot spot residues computationally. Some of these methods work as servers such as Robetta (10,11) and KFC server (23). Robetta server (10,11) performs computational alanine scanning based on estimating energy (including van der Waals, H-bonds) at atomic level for a given complex and outputs changes in the binding free energy values for each residue in the interface. KFC server (23) predicts hot spots for a given complex using a machine learning approach which considers the shape specificity and surrounding structural features of the residues. Server output is the confidence scores and the predictions. Results can be visualized by an interactive viewer.
Here, we present HotPoint web server, which provides a user-friendly interface to run the method developed by Tuncbag et al. (19) for online prediction of hot spots in protein interfaces. Our aim is to provide an efficient server at a single location for analysis of any protein–protein interface which can be utilized by researchers interested in protein binding sites. The method principally considers the solvent accessibility and the total contact potential of the interface residues. The output tabulates the interface residues with the highlighted hot spots and their features. Additionally, it provides an interactive 3D visualization of the submitted protein–protein interface with the predicted hot spots for observing their localization. Distinct features of HotPoint from existing servers (Robetta and KFC server) are the improved efficiency and accuracy. The calculation of solvent accessibility and pair potentials of residues are faster than atomic level computations performed by Robetta, and the prediction accuracy is higher than both Robetta and KFC server.
THE HOTPOINT METHOD
HotPoint is based on a few simple rules consisting of solvent accessibility and energetic contribution of residues. The thresholds of the model are adjusted according to a data set composed of 150 experimentally alanine mutated residues of which 58 residues are hot spots and 92 residues are non-hot spots. The interface residues, whose mutations change the binding free energy at least 2.0 kcal/mol, are considered as experimental hot spots. If the mutation results in a change <0.4 kcal/mol, that residue is labeled as experimental non-hot spot. The independent test set is derived from Binding Interface Database (BID) (7), composed of 112 residues (of which 54 residues are hot spots and 58 residues are non-hot spots). The predictive performance of this method is assessed using accuracy (the ratio of number of correctly predicted residues to number of all predicted residues), precision (the ratio of number of correctly classified hot spot residues to the number of all residues classified as hot spots), recall (the proportion of number of correctly classified hot spot residues to the number of all hot spot residues), specificity (the proportion of number of correctly predicted non-hot spot residues to the number of all non-hot spot residues) and F1 score (the balance between precision and recall).
Several empirical and machine learning methods are trained and tested using several features [relative accessible surface area (ASA) in complex state, relative change in ASA upon complexation, conservation, amino acid propensity and total contact potential]. At the end, the best performance is achieved by an empirical model based on relative accessibility in complex state and total pair potentials. According to this model, if an individual interface residue is buried (its relative ASA in complex state is ≤20%) and its total contact potential is ≥18.0, this residue is flagged as a hot spot; else, it is flagged as a non-hot spot. The thresholds of the model (20% and 18.0) are inferred from training set. This model demonstrates an accuracy of 0.70, a precision of 0.73, a recall of 0.59 and a specificity of 0.79 on the independent test set, which exceeds the performance of existing approaches [such as, KFC (15), KFCA (15), ISIS (18), Robetta (11)] and machine learning approaches (such as, SVM, BayesNet, decision tree, etc.) on the same test set. The details of the data sets, methodology and an exhaustive comparison with other approaches are available in Tuncbag et al. (19).
HOTPOINT WEB SERVER
The HotPoint web server is available at http://prism.ccbb.ku.edu.tr/hotpoint. Server interface is coded in PHP. The code to predict hot spots is written in Python.
Input
Input data is the protein structure in PDB formatted coordinate file, two chain identifiers forming the interface and the interface definition. User can either run the server with default distance thresholds to extract interface residues or can change the interface definition by submitting a distance threshold. There are two options to submit a structure file. User can enter the four letter PDB code of a protein which is directly downloaded from the ftp site of PDB. The second option is uploading a structure file that is in the PDB format. HotPoint requires two chain identifiers which confine to a protein interface. Server does not work for PDB files containing only one chain and returns an error. For NMR structures, it uses the first model in the prediction and gives results for the first model. HotPoint is specific to protein–protein interfaces; chains corresponding to DNA structures return a warning in the web server.
When there is not enough input data, the server informs the users of what is missing. The HotPoint web server is free and open to all users and there are no login requirements.
Extraction of computational hot spots
When a protein structure with its chain identifiers is submitted, HotPoint server starts the calculation of three consecutive steps:
Extraction of interface residues: a protein interface is defined as a set of amino acids which represents a region that links two protein chains by non-covalent interactions. According to the default interface definition in the server, if the distance between any two atoms belonging to two residues, one from each chain, is less than the sum of their van der Waals radii plus a 0.5 Å tolerance, these two residues are defined as interacting. Users can change this definition by submitting a distance threshold.
Calculation of the features: Residue solvent accessibilities are calculated using Naccess (24). The residue accessibilities in complex state and in monomer state are converted into relative accessibilities by dividing them to maximum accessibility of that residue. Knowledge-based solvent mediated inter-residue potentials are taken from Keskin et al. (25). In the contact potential matrix, there are 210 distinct contact potentials between all possible pairs of 20 amino acids in RT unit (R, universal gas constant; T, absolute temperature). To calculate the total contact potential of a residue in the interface, server extracts the neighbors of that residue whose side chain center of mass are closer than the cutoff (7.0 Å). Another constraint for neighbor extraction is that they should not be close neighbor in sequence (|i−j| ≥4 where i and j are residue numbers). Total contact potential of the residue is defined as the absolute of the sum of the contact potentials with its neighbors (19).
Prediction based on empirical model: Finally, the empirical model [presented in Tuncbag et al. (19)] is applied on the residue to determine whether it is a computational hot spot or not. If the relative accessibility of an individual interface residue is ≤20% and its total contact potential is ≥18.0, it is labeled as hot spot (19).
Output
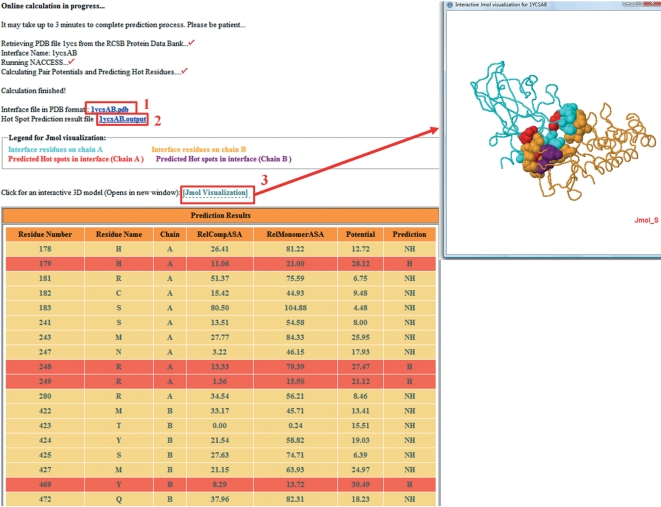
During the processing, the server informs users about the steps it is performing. The output of the server is a table consisting of the interface residues with their features (Figure 1). The interface residues are tabulated with chain names, one-letter residue names, residue numbers, their relative ASA in complex, relative ASA in monomer and total pair potentials. In the last column of the table, the prediction is presented as H (hot spot) or NH (non-hot spot). Background of the predicted hot spots is highlighted with red color. The prediction results as a text file and interface residue coordinates in PDB file format are also downloadable by the user. In this way, the results can be visualized in any visualization tool. Besides the downloadable files, overall complex, the interface residues and hot spots can be visualized interactively using the Jmol (26) applet window in the HotPoint server.
Figure 1.
The output page of HotPoint for the p53 DNA binding domain/53BP2 protein complex (pdb:1ycs, chain A and B). Interface residues of this complex are shown in the table with hot spot predictions: (1) the coordinates of interface residues can be downloaded; (2) hot spot prediction results are also downloadable; and (3) interface with predicted hot spots can be visualized by JMol.
An independent case study: Interleukin-2 and its receptor complex
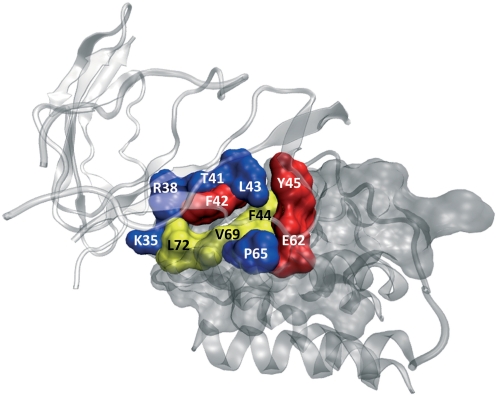
Interleukin-2 (IL-2) is a cytokine immune system signaling molecule. IL-2 gets functional when it associates with the IL-2 receptor. To find the residues necessary for binding, several residues (K35, R38, M39, T41, F42, K43, F44, Y45, E62, P65, V69 and L72) on IL-2 are mutated to alanine. Among these residues, F42, Y45 and E62 reduce binding affinity of IL-2 to its receptor >100-folds. Further, small inhibitor molecule SP4206 also targets these hot spots of the receptor (27). HotPoint predicts all three experimental hot spots (F42, Y45 and E62) correctly for the IL-2/IL-2 receptor complex (PDB code: 1z92, chain A is IL-2 and chain B is IL-2 receptor). According to our interface definition, M39 cannot be found in the interface residues. So, for the remaining eight residues, HotPoint labels five residues (K35, R38, T41, K43 and P65) as non-hot spot, correctly. However, three residues come as false positives (F44, V69 and L72) from HotPoint prediction. As a result, 8 out of 11 alanine mutations are correctly predicted. This protein complex is independent from the training and test sets. The predictions are illustrated in Figure 2 in 3D using the output files obtained from HotPoint.
Figure 2.
IL-2/IL-2 receptor complex. The PDB code for this complex is 1z92. The red-colored residues are correctly predicted hot spots. The blue-colored ones are correctly predicted non-hot spots. The yellow-colored residues represent non-hot spot residues that are incorrectly predicted as hot spots.
CONCLUSIONS
A small subset of residues in protein interfaces comprises a large portion of binding free energy, namely hot spots. We present HotPoint server to determine computational hot spots in protein interfaces based on solvent accessibility and pair potentials which allows online calculation for all protein interfaces within practical running times. Further, the model outperforms other existing approaches. It tabulates residue level features and prediction results for a given protein complex which are also downloadable. We hope that with its simple architecture and visualization tool, HotPoint would be useful both for the experimentalists and computational scientist working on protein recognition, modeling of protein complexes and drug design.
FUNDING
TUBITAK (Research Grant No 109T343 and 109E207). TUBITAK fellowship (to N.T.). Funding for open access charge: Koc University.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We thank to Abdullah M. Turan for his help in developing the web service.
REFERENCES
- 1.Bogan AA, Thorn KS. Anatomy of hot spots in protein interfaces. J. Mol. Biol. 1998;280:1–9. doi: 10.1006/jmbi.1998.1843. [DOI] [PubMed] [Google Scholar]
- 2.Clackson T, Wells JA. A hot spot of binding energy in a hormone-receptor interface. Science. 1995;267:383–386. doi: 10.1126/science.7529940. [DOI] [PubMed] [Google Scholar]
- 3.Wells JA. Systematic mutational analyses of protein-protein interfaces. Meth. Enzymol. 1991;202:390–411. doi: 10.1016/0076-6879(91)02020-a. [DOI] [PubMed] [Google Scholar]
- 4.Keskin O, Ma B, Nussinov R. Hot regions in protein–protein interactions: the organization and contribution of structurally conserved hot spot residues. J. Mol. Biol. 2005;345:1281–1294. doi: 10.1016/j.jmb.2004.10.077. [DOI] [PubMed] [Google Scholar]
- 5.Keskin O, Gursoy A, Ma B, Nussinov R. Principles of protein–protein interactions: what are the preferred ways for proteins to interact? Chem. Rev. 2008;108:1225–1244. doi: 10.1021/cr040409x. [DOI] [PubMed] [Google Scholar]
- 6.Keskin O, Tuncbag N, Gursoy A. Characterization and prediction of protein interfaces to infer protein-protein interaction networks. Curr. Pharm. Biotechnol. 2008;9:67–76. doi: 10.2174/138920108783955191. [DOI] [PubMed] [Google Scholar]
- 7.Fischer TB, Arunachalam KV, Bailey D, Mangual V, Bakhru S, Russo R, Huang D, Paczkowski M, Lalchandani V, Ramachandra C, et al. The binding interface database (BID): a compilation of amino acid hot spots in protein interfaces. Bioinformatics. 2003;19:1453–1454. doi: 10.1093/bioinformatics/btg163. [DOI] [PubMed] [Google Scholar]
- 8.Thorn KS, Bogan AA. ASEdb: a database of alanine mutations and their effects on the free energy of binding in protein interactions. Bioinformatics. 2001;17:284–285. doi: 10.1093/bioinformatics/17.3.284. [DOI] [PubMed] [Google Scholar]
- 9.Guerois R, Nielsen JE, Serrano L. Predicting changes in the stability of proteins and protein complexes: a study of more than 1000 mutations. J. Mol. Biol. 2002;320:369–387. doi: 10.1016/S0022-2836(02)00442-4. [DOI] [PubMed] [Google Scholar]
- 10.Kortemme T, Baker D. A simple physical model for binding energy hot spots in protein-protein complexes. Proc. Natl Acad. Sci. USA. 2002;99:14116–14121. doi: 10.1073/pnas.202485799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kortemme T, Kim DE, Baker D. Computational alanine scanning of protein-protein interfaces. Sci STKE. 2004;2004:pl2. doi: 10.1126/stke.2192004pl2. [DOI] [PubMed] [Google Scholar]
- 12.Guharoy M, Chakrabarti P. Empirical estimation of the energetic contribution of individual interface residues in structures of protein-protein complexes. J. Comput. Aided Mol. Des. 2009;23:645–654. doi: 10.1007/s10822-009-9282-3. [DOI] [PubMed] [Google Scholar]
- 13.Assi SA, Tanaka T, Rabbitts TH, Fernandez-Fuentes N. PCRPi: Presaging Critical Residues in Protein interfaces, a new computational tool to chart hot spots in protein interfaces. Nucleic Acids Res. 2009;38:e86. doi: 10.1093/nar/gkp1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cho KI, Kim D, Lee D. A feature-based approach to modeling protein-protein interaction hot spots. Nucleic Acids Res. 2009;37:2672–2687. doi: 10.1093/nar/gkp132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Darnell SJ, Page D, Mitchell JC. An automated decision-tree approach to predicting protein interaction hot spots. Proteins. 2007;68:813–823. doi: 10.1002/prot.21474. [DOI] [PubMed] [Google Scholar]
- 16.Guney E, Tuncbag N, Keskin O, Gursoy A. HotSprint: database of computational hot spots in protein interfaces. Nucleic Acids Res. 2008;36:D662–D666. doi: 10.1093/nar/gkm813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lise S, Archambeau C, Pontil M, Jones DT. Prediction of hot spot residues at protein-protein interfaces by combining machine learning and energy-based methods. BMC Bioinformatics. 2009;10:365. doi: 10.1186/1471-2105-10-365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ofran Y, Rost B. Protein-protein interaction hotspots carved into sequences. PLoS Comput. Biol. 2007;3:e119. doi: 10.1371/journal.pcbi.0030119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tuncbag N, Gursoy A, Keskin O. Identification of computational hot spots in protein interfaces: combining solvent accessibility and inter-residue potentials improves the accuracy. Bioinformatics. 2009;25:1513–1520. doi: 10.1093/bioinformatics/btp240. [DOI] [PubMed] [Google Scholar]
- 20.Gonzalez-Ruiz D, Gohlke H. Targeting protein-protein interactions with small molecules: challenges and perspectives for computational binding epitope detection and ligand finding. Curr. Med. Chem. 2006;13:2607–2625. doi: 10.2174/092986706778201530. [DOI] [PubMed] [Google Scholar]
- 21.Huo S, Massova I, Kollman PA. Computational alanine scanning of the 1:1 human growth hormone-receptor complex. J. Comput. Chem. 2002;23:15–27. doi: 10.1002/jcc.1153. [DOI] [PubMed] [Google Scholar]
- 22.Rajamani D, Thiel S, Vajda S, Camacho CJ. Anchor residues in protein-protein interactions. Proc. Natl Acad. Sci. USA. 2004;101:11287–11292. doi: 10.1073/pnas.0401942101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Darnell SJ, LeGault L, Mitchell JC. KFC Server: interactive forecasting of protein interaction hot spots. Nucleic Acids Res. 2008;36:W265–W269. doi: 10.1093/nar/gkn346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hubbard SJ, Thornton JM. Department of Biochemistry and Molecular Biology. University College: London; 1993. [Google Scholar]
- 25.Keskin O, Bahar I, Badretdinov AY, Ptitsyn OB, Jernigan RL. Empirical solvent-mediated potentials hold for both intra-molecular and inter-molecular inter-residue interactions. Protein Sci. 1998;7:2578–2586. doi: 10.1002/pro.5560071211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Herraez A. Biomolecules in the computer: Jmol to the rescue. Biochem Mol Biol Educ. 2006;34:255–261. doi: 10.1002/bmb.2006.494034042644. [DOI] [PubMed] [Google Scholar]
- 27.Thanos CD, DeLano WL, Wells JA. Hot-spot mimicry of a cytokine receptor by a small molecule. Proc. Natl Acad. Sci. USA. 2006;103:15422–15427. doi: 10.1073/pnas.0607058103. [DOI] [PMC free article] [PubMed] [Google Scholar]