Abstract
MAGIA (miRNA and genes integrated analysis) is a novel web tool for the integrative analysis of target predictions, miRNA and gene expression data. MAGIA is divided into two parts: the query section allows the user to retrieve and browse updated miRNA target predictions computed with a number of different algorithms (PITA, miRanda and Target Scan) and Boolean combinations thereof. The analysis section comprises a multistep procedure for (i) direct integration through different functional measures (parametric and non-parametric correlation indexes, a variational Bayesian model, mutual information and a meta-analysis approach based on P-value combination) of mRNA and miRNA expression data, (ii) construction of bipartite regulatory network of the best miRNA and mRNA putative interactions and (iii) retrieval of information available in several public databases of genes, miRNAs and diseases and via scientific literature text-mining. MAGIA is freely available for Academic users at http://gencomp.bio.unipd.it/magia.
INTRODUCTION
MicroRNAs (miRNAs) are small non-coding RNAs acting as post-transcriptional regulators of gene expression whose discovery added a novel layer of genetic regulation in a wide range of biological processes, including cell differentiation, organogenesis and development (1–3). Deregulation of miRNAs expression plays a critical role in the pathogenesis of genetic and multifactorial disorders, as well as most human cancers (4). By imperfect base pairing with the 3′-untranslated region (3′-UTR) of their target mRNAs, mature miRNAs can cause translation inhibition or mRNA cleavage, depending on the degree of complementarity between the miRNA and its target sequence (5,6). Given that miRNAs can have multiple targets and that each protein-coding gene can be targeted by multiple miRNAs, it has been suggested that more than one third of human genes could be regulated by miRNAs. In this perspective, the networks of post-transcriptional regulatory relationships tend to have a highly complex nature (7).
Among the computational approaches applied to predict miRNA targets could be found (i) algorithms based on sequence search similarity, possibly considering target site evolutionary conservation [miRanda (8), TargetScan (9) and PicTar (10)] and (ii) algorithms based on thermodynamic stability of the RNA–RNA duplex, considering free energy minimization [RNAhybrid (11) and PITA (12)]. However, all available software is plagued by a significant fraction of false positives. This is caused not only by the limited comprehension of the molecular basis of miRNA–target pairing, but also by the context-dependency of post-transcriptional regulation. According to the increasing experimental evidences supporting the miRNA mechanism of target degradation rather than translational repression, the integration of target predictions with miRNA and gene expression profiles has been proposed to improve the detection of functional miRNA–mRNA relationships. Since miRNAs tend to down-regulate target mRNAs (13–15), the expression profiles of genuinely interacting pairs are expected to be anti-correlated. Integrative analysis can be performed adopting a variational Bayesian model (16,17), or by using a non-heuristic methodology based on the anti-correlation between miRNA and mRNA expression profiles.
Unfortunately, the combination of large-scale target prediction results obtained with different algorithms is not straightforward for most experimental researchers, whereas the integrative analysis of miRNA and gene expression profiles is complicated by the many-to-many nature of predicted relationships and target annotations to be considered.
Recently, two web tools have been introduced to enhance the functional insights of target predictions: miRGator (18) and DIANA-microT web servers (19). They may help the elucidation of biological processes, functions and pathways targeted by miRNAs through the integration of target predictions with information from different gene, protein and functional annotation databases.
Mirz (20) integrates the smiRNAdb miRNA expression atlas, based on small mammalian RNA library sequencing, and the ElMMo miRNA target prediction algorithm, allowing the user to restrict the target prediction to specific miRNAs, selected by expression characteristics.
Finally, the web tool MMIA [miRNA and mRNA Integrated Analysis (21)], integrates miRNAs and mRNA expression data using only significantly up- and down-regulated features without taking into account their whole expression profile, losing, in this way, a key information for the calculation of the expression anti-correlation degree. Available tools are definitely not adequate to the rapidly increasing amount of matched miRNA–gene profiles (miRNA and gene expression profiles quantified on exactly the same set of biological samples), the analysis of which could gain a remarkable advantage from target predictions and miRNA–gene expression profiles integration.
Here we present MAGIA (miRNA and genes integrated analysis, freely available at http://gencomp.bio.unipd.it/magia), a novel web tool that allows to integrate target predictions and gene expression profiles using different relatedness measures either for matched or un-matched expression profiles, using miRNA–mRNA bipartite networks reconstruction, gene functional enrichment and pathway annotations for results browsing.
DESCRIPTION OF THE TOOL
MAGIA outline
MAGIA is a novel web-based tool that allows (i) to retrieve and browse updated miRNA target predictions for human miRNAs, based on a number of different algorithms (PITA, miRanda and TargetScan), with the possibility of combining them with Boolean operators, (ii) the direct integration through different functional measures (parametric and non-parametric correlation indexes, a variational Bayesian model, mutual information and a meta-analysis approach based on P-value combination) of mRNA and miRNA expression data (iii) the construction of bipartite regulatory networks of the best miRNAs and mRNA putative interaction and finally and (iv) to retrieve information available in several public databases of genes, miRNAs and diseases and via scientific literature text-mining. Step-by-step tutorial pages and sample data sets are provided to the user to easily introduce him to the use of the tool.
MAGIA is divided into two separate sections: the query and the analyses frameworks whose aims are described in the following paragraphs.
Query
The query section of MAGIA allows the user to search for target predictions of specific miRNAs obtained through PITA, miRanda or TargetScan or combinations thereof, setting cutoffs on prediction scores. Target prediction algorithms have been selected according to their different strategies: sequence similarity (miRanda), sequence similarity with conservation (TargetScan) and sequence similarity with free energy minimization (PITA). We run each of these algorithms on our servers to update predictions every 6 months. The query output is a table including, for all considered miRNAs, the list of predicted target genes or transcripts with the different prediction scores according to the method(s) chosen by the user. The same information may be downloaded as a text file for processing and further elaboration.
Analysis
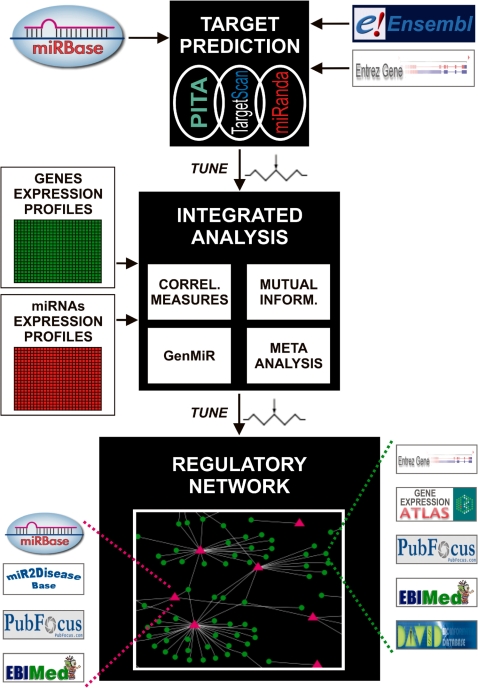
The analysis pipeline is composed by three different steps through which MAGIA refines target predictions using miRNA and mRNA gene expression data (Figure 1): (i) selection of the organism, the gene or transcript annotation (EntrezGene, RefSeq, ENSEMBL gene or transcript) and of the integration method or the relatedness measure; (ii) choice of target prediction algorithms, their score cut-offs and Boolean combinations and (iii) upload of two matrices representing mRNA and miRNA normalized expression profiles. MAGIA takes into account two different experimental designs: (i) mRNA and miRNA data collected on different biological samples, resulting in different sample sizes (hereafter called non-matched case) and (ii) mRNA and miRNA expression data obtained from the same biological samples (the matched case). The tool employs a meta-analysis approach based on a P-value combination in the first case, while one of four different measures of relatedness can be adopted for the analysis of matched profiles: Spearman and Pearson correlation, mutual information, and a variational Bayesian model. Computational intensive calculations of MAGIA analyses are carried out by a multicore cluster.
Figure 1.
Scheme of the three main steps of analysis pipeline performed by MAGIA web tool.
Input files
The third step of the MAGIA analysis pipeline takes as input two expression matrices (in the tab-delimited format) with genes and miRNAs on the rows and samples on the columns. When profiles are matched, the names of the columns of mRNA and miRNA data sets should correspond exactly, while in the non-matched-case the columns labels should represent sample classes: samples belonging to the same class should have the same label. The first column of both matrices should represent miRNA and gene IDs. MAGIA allows EntrezGene or Ensembl IDs for genes and RefSeq or Ensembl IDs for transcripts, while miRNA IDs must represent miRBase-compliant mature miRNA identifiers. Expression matrices should be pre-processed and a filtering procedure for the removal of invariable (‘flat’) expression profiles is highly recommended. A series of quality checks are performed during the upload.
Sample files for miRNA and gene expression, fully compliant with the user choices of steps 1 and 2 are also provided in this step, for tutorial purposes. These sample files derive from expression data publicly available at GEO database (GSE14834) (22).
Target predictions
We have used the miRanda and PITA algorithms to compute miRNA target predictions over up-to-date versions 56 and 38 of ENSEMBL and RefSeq transcript sequences, respectively. The miRNA sequences were downloaded from mirBase version 14. Based on known transcript to gene correspondences, gene-centered predictions were then derived combining transcript-based results into a single group for each gene. In this way a gene is predicted target of a given miRNA if at least one of its transcripts carries predicted target site(s). TargetScan predictions (version 5.0) were downloaded from http://www.targetscan.org.
Computation of interaction measures
Pearson and Spearman correlations
Correlation indicates the strength and direction of a linear relationship between two random variables. Parametric (Pearson) or non-parametric (Spearman) correlation coefficients are computed in the case of matched samples between gene/transcript and miRNA data. In general, non-parametric statistic has different expected values from the Pearson correlation coefficient, even for large samples. Since they estimate different population parameters, they cannot be directly compared: they generally should be viewed as alternative measures of association. The non-parametric coefficient should be chosen in case of outliers or with small number of measures; otherwise a parametric approach may be more appropriate. Moreover, Pearson coefficient testing requires that both variables derive from a bivariate normal distribution, an assumption not necessary for the Spearman coefficient testing.
The tool computes correlation coefficients for all the predicted miRNA–target interactions and also provides a false discovery rate (FDR, following Benjamini and Hochberg estimation method) for each one.
Mutual information
Mutual information is a measure of the mutual dependence of two variables. Intuitively, it captures the information that a variable X (a gene expression profile) and a variable Y (a miRNA expression profile) share: how much the knowledge of one of these variables reduces our uncertainty about the other. Thus, the mutual information can be interpreted as a generalized measure of correlation, analogous to the Pearson correlation, but sensitive to any functional relationship, not just to linear dependencies. There are several possible strategies for the reliable estimation of the mutual information in case of finite data, each of them characterized by a systematic error due to the finite size sample [see (23) for a review]. In particular, following the Kraskov and colleagues (2003) approach (24), MAGIA calculates mutual information based on nearest neighbor distances with k = 5. Mutual information, identifying any functional relationship between miRNA and gene expression profiles, does not allow the identification of the sign of such relationship.
GenMir++
The variational Bayesian model, called GenMiR++ (16,17) uses as prior information target predictions derived from one of the previous mentioned algorithms (e.g. PITA) and updates such information using expression matrices. It combines predictions with miRNA and mRNA expression profiles, under the assumption of anti-correlation. Under a complex model, the posterior probability of miRNA–gene interactions (S) is calculated, known the target predictions (C), expression matrices (X and Z), by integrating over nuisance variables gamma (Γ, tissue scaling) and lambda (Λ, regulatory weights) and other parameters in the equation,
An estimate of such posterior probability is calculated through an EM algorithm. Thus GenMir++ could have convergence problems, particularly in case of non-sparse incidence matrices.
Meta-analysis
The meta-analysis approach is suggested only in the case of non-matched biological samples. Given the diverse nature and number of samples between miRNA and gene profiles, neither correlation coefficients nor mutual information or posterior probabilities can be computed. MAGIA adopts in their place a meta-analyses approach based on P-value combination allowing, unlike other web tools, the presence of more than two groups. Empirical Bayes test (25) (as implemented in limma package in R) is separately performed on miRNA and mRNA expression levels and lists of differentially miRNAs and genes are stored. Then, only for predicted miRNA–mRNA interactions (based on the target prediction algorithms the user has chosen) the inverse Chi-squared approach (26) is used to combine miRNAs and genes P-values. In particular, in the case of a two classes experimental design, P-values of over-expressed miRNAs (e.g. under-expressed in Class 1 versus Class 2) are combined with those of under-expressed genes (Class 1 versus Class 2) and vice versa. In the case of more than two classes the tool combines P-values derived from miRNAs, genes and from the test on Spearman correlation coefficient computed between vectors representing the average expression values of miRNAs and genes within each class. Only the interactions with small P-values (<0.1) will be considered as functional.
Output and links to other database resources
MAGIA reports results in a web page containing different sections. For the top 250 most probable functional miRNA–mRNA interactions according to the association measure selected by the user, the interactive bipartite regulatory network obtained through the analysis is reported along with the corresponding browsable table of relationships. It gives a hyperlink allowing the functional enrichment analysis by the DAVID web tool (27) on the desired number of target genes. The tool also provides the complete list of the predicted interactions, ranked by statistical significance computed from the integrated expression data analysis. Such information is given as HTML tables and as two (Cytoscape-compliant) flat files for network reconstruction. Each mRNA, miRNA or miRNA–mRNA interaction can be further investigated by the user and used for different queries. In particular, each gene is linked to EntrezGene (28), and ArrayExpress Atlas (29) databases, each miRNA is linked to miRNA2disease (30) and miRecords (31). Furthermore, to allow efficient and systematic retrieval of statements from Medline, MAGIA directly links results to PubFocus (32) and EbiMEd (33) for a text-mining search using genes and miRNAs as keywords.
Bi-partite networks visualizations
The miRNA and gene bipartite network is rendered using Graphviz (http://www.graphviz.org/) open source graph visualization software. Each node of the network can be selected and the user is directly linked to the corresponding miRNA/gene full interactions results. Thus the user is allowed to ‘walk through the network’ following miRNA and gene interactions. The complete list of significant interactions can be downloaded as a tab-delimited text file that can be imported into Excel or Cytoscape, to allow further processing.
Case study
As a benchmark case study we used the mRNA and miRNA expression profiles published by Fulci et al. (22) and publicly available in the GEO database (GSE14834). In this study, the Authors investigated miRNA and gene expression profiles in a series of adult Acute Lymphoblastic Leukemia (ALL) cases. ALL is a heterogeneous disease comprising several subentities that differ for both immunophenotypic and molecular characteristics. In particular, T-lineage and B-lineage harboring specific molecular lesions have been considered by expression analyses.
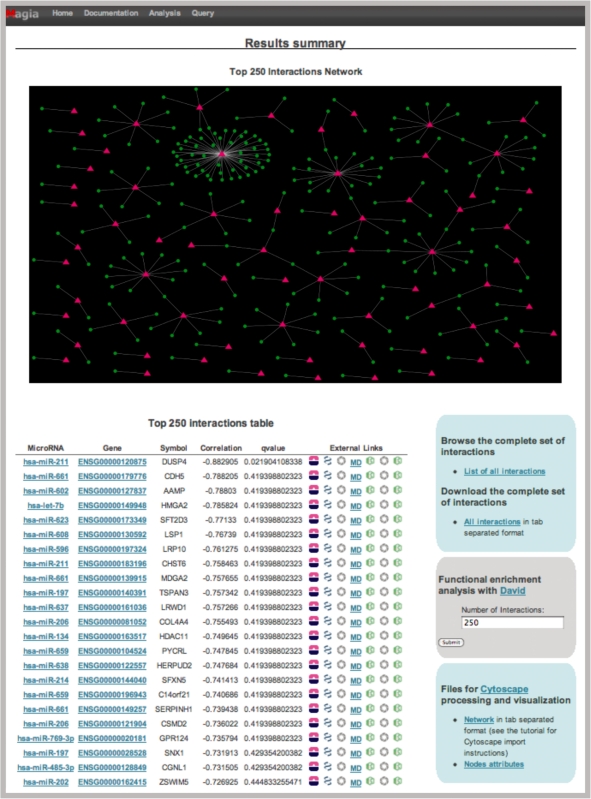
In this example, we choose EntrezGene IDs, Pearson correlation measure and the intersection of TargetScan and PITA target prediction algorithms. A total number of 468 miRNA–mRNA interactions with absolute correlations >0.25 have been identified, 249 of these show negative while 219 show positive correlation coefficients. Among the 468 putative interactions 23 have an FDR value <0.1. Figure 2 shows, for the top 250 miRNA–target relationships most supported by expression data, the bipartite network and the corresponding list with hyperlinks to mirBase, EntrezGene, PubFocus, EbiMed and mir2disease, whereas for all predicted interactions, a link to an html table and to a tab delimited flat file Cytoscape compliant are given, as well as the link to the DAVID annotation tool for a number of interactions that can be defined by the user (default is set to 250).
Figure 2.
Screenshot of the MAGIA analysis results summary page, obtained with the ALL data case study analysis. The summary page reports the regulatory network corresponding to the 250 relations most supported by expression data and the corresponding details, as genes and miRNAs involved, with links to databases and text-mining tools. This is also the entry point to reach gene- or miRNA-centered pages, to carry out functional enrichment analysis and to download complete results.
In this example, the top 250 interactions include a total number of 81 different miRNAs and of 197 different genes. Pathways enrichment analysis, conducted on target genes and aiming at clarifying the role of miRNAs in terms of cell activities under post-transcriptional regulation, leads to highly relevant and interesting results: chronic myeloid leukemia is the KEGG most enriched pathway according to DAVID, followed by Wnt-signaling pathway, pancreatic cancer and ubiquitin mediated proteolysis. Chronic myelogenous leukemia is a biphasic disease, initiated by expression of the BCR/ABL fusion gene product in self-renewing, hematopoietic stem cells; among the 43 B-ALL patients used in the expression analysis 17 had a BCR/ABL rearrangement. On the other hand, the Wnt family of secreted glycoproteins regulate early B cell growth and survival (34) and aberrant activation of the Wnt-signaling pathway has major oncogenic effects (35). Finally, the ubiquitin pathway plays a central role in the regulation of cell growth and cell proliferation controlling the abundance of key cell-cycle proteins. Increasing evidence indicates that unscheduled proteolysis of many cell-cycle regulators contributes significantly to tumorigenesis and is indeed found in many types of human cancers (36).
Among the top miRNA–gene anti-correlated interactions we found RALB (v-ral simian leukemia viral oncogene homolog B), a gene encoding a GTP-binding protein that belongs to the small GTPase superfamily and Ras family of proteins, highly associated to either let-7d (r = −0.82) and let-7c (r = −0.71). Recently RALA and RALB have shown to collaborate to maintain tumorigenicity through regulation of both proliferation and survival (37) while both let-7d and let-7c have been shown to be involved in the human acute promyelocytic leukemia (38).
hsa-miR-222 and let-7e have been recently found to be two of the most discriminant miRNAs markers between ALL and AML (39) and in our analysis have been found highly anti-correlated with respectively ETS1 (v-ets erythroblastosis virus E26 oncogene homolog 1) (r = −0.58) recently found to be involved in tumor development and progression (40) and with p53 (r = –0.51) whose oncogenic role has been extensively studied in the last years (41).
Several other interactions have been reported by MAGIA, most of them including miRNAs and/or genes involved in tumor development and progression. Indeed, repeating the sample analysis with the same expression data and settings indicated above, but only for the 12 miRNAs reportedly differentially expressed across samples (22) an interaction biologically relevant and validated (according to Diana Tarbase and miRecords), regarding hsa-let-7e and HMGA2 (high mobility group AT-hook 2 gene) is indicated by MAGIA at the first ranked position. While a complete investigation of biological relevance of all interactions reported by MAGIA is beyond the scope of this work, they validate the MAGIA integrative approach, the usefulness of the display of results and the discovery power of data analysis with this tool.
CONCLUSIONS
The integrative analysis of target prediction, miRNA and gene expression profiles is not straightforward for most experimental researchers, not only for problems regarding miRNA and targets annotations, but also for the many-to-many nature of predicted relationships to be considered and the extensive time requirements of computations. However, there is an increasing amount of experimental studies aiming at gaining molecular understanding of biological processes or diseases from the computation and the visualization of high-throughput systems biology analyses results. Available tools are not adequate to the rapidly increasing amount of matched miRNA–gene profiles, the analysis of which could gain a remarkable advantage from target predictions and miRNA–gene expression profiles integration. MAGIA (MiRNA And Genes Integrated Analysis) tries to fill these gaps allowing the combination of target predictions for either matched or un-matched expression miRNA–gene profiles. Using different relatedness measures and integration methods, MAGIA refines target predictions and reconstructs miRNA–gene bipartite networks. In this context, MAGIA is a useful, timely and easy-to-use web tool that will facilitate users in the investigation of the post-transcriptional regulatory networks and in the discovery of biologically relevant regulatory circuits.
FUNDING
Fondazione Cassa di Risparmio di Padova e Rovigo (Progetti Eccellenza 2006); University of Padova (60A06-1977/09, 60A06-8282/09, 60A06-0972/10 and 60A06-4898/10); Italian Ministry for University and Research (PRIN 2007CHSMEB_002); AIRC (Italian Association for Cancer Research; Regional Research Program 2008). Funding for open access charges: University of Padova.
Conflict of interest statement. None declared.
REFERENCES
- 1.Zhao Y, Srivastava D. A developmental view of microRNA function. Trends Biochem. Sci. 2007;32:189–197. doi: 10.1016/j.tibs.2007.02.006. [DOI] [PubMed] [Google Scholar]
- 2.Fazi F, Nervi C. MicroRNA: basic mechanisms and transcriptional regulatory networks for cell fate determination. Cardiovasc. Res. 2008;79:553–561. doi: 10.1093/cvr/cvn151. [DOI] [PubMed] [Google Scholar]
- 3.Callis TE, Wang DZ. Taking microRNAs to heart. Trends Mol. Med. 2008;14:254–260. doi: 10.1016/j.molmed.2008.03.006. [DOI] [PubMed] [Google Scholar]
- 4.Calin GA, Ferracin M, Cimmino A, Di Leva G, Shimizu M, Wojcik SE, Iorio MV, Visone R, Sever NI, Fabbri M, et al. A MicroRNA signature associated with prognosis and progression in chronic lymphocytic leukemia. N Eng. J. Med. 2005;353:1793–1801. doi: 10.1056/NEJMoa050995. [DOI] [PubMed] [Google Scholar]
- 5.Rajewsky N. microRNA target predictions in animals. Nat. Genet. 2006;38(Suppl.):S8–S13. doi: 10.1038/ng1798. [DOI] [PubMed] [Google Scholar]
- 6.Flynt AS, Lai EC. Biological principles of microRNA-mediated regulation: shared themes amid diversity. Nat. Rev. Genet. 2008;9:831–842. doi: 10.1038/nrg2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brown JR, Sanseau P. A computational view of microRNAs and their targets. Drug Discov. Today. 2005;10:595–601. doi: 10.1016/S1359-6446(05)03399-4. [DOI] [PubMed] [Google Scholar]
- 8.Enright AJ, John B, Gaul U, Tuschl T, Sander C, Marks DS. MicroRNA targets in Drosophila. Genome Biol. 2003;5:R1. doi: 10.1186/gb-2003-5-1-r1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB. Prediction of mammalian microRNA targets. Cell. 2003;115:787–798. doi: 10.1016/s0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- 10.Krek A, Grun D, Poy MN, Wolf R, Rosenberg L, Epstein EJ, MacMenamin P, da Piedade I, Gunsalus KC, Stoffel M, et al. Combinatorial microRNA target predictions. Nature Genet. 2005;37:495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- 11.Rehmsmeier M, Steffen P, Hochsmann M, Giegerich R. Fast and effective prediction of microRNA/target duplexes. RNA. 2004;10:1507–1517. doi: 10.1261/rna.5248604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kertesz M, Iovino N, Unnerstall U, Gaul U, Segal E. The role of site accessibility in microRNA target recognition. Nat. Genet. 2007;39:1278–1284. doi: 10.1038/ng2135. [DOI] [PubMed] [Google Scholar]
- 13.Bagga S, Bracht J, Hunter S, Massirer K, Holtz J, Eachus R, Pasquinelli AE. Regulation by let-7 and lin-4 miRNAs results in target mRNA degradation. Cell. 2005;122:553–563. doi: 10.1016/j.cell.2005.07.031. [DOI] [PubMed] [Google Scholar]
- 14.Lim LP, Lau NC, Garrett-Engele P, Grimson A, Schelter JM, Castle J, Bartel DP, Linsley PS, Johnson JM. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- 15.Wu L, Belasco JG. Let me count the ways: mechanisms of gene regulation by miRNAs and siRNAs. Mol. Cell. 2008;29:1–7. doi: 10.1016/j.molcel.2007.12.010. [DOI] [PubMed] [Google Scholar]
- 16.Huang JC, Babak T, Corson TW, Chua G, Khan S, Gallie BL, Hughes TR, Blencowe BJ, Frey BJ, Morris QD. Using expression profiling data to identify human microRNA targets. Nat. Methods. 2007;4:1045–1049. doi: 10.1038/nmeth1130. [DOI] [PubMed] [Google Scholar]
- 17.Huang JC, Morris QD, Frey BJ. Bayesian inference of MicroRNA targets from sequence and expression data. J. Comput. Biol.: J. Comput. Mol. Cell Biol. 2007;14:550–563. doi: 10.1089/cmb.2007.R002. [DOI] [PubMed] [Google Scholar]
- 18.Nam S, Kim B, Shin S, Lee S. miRGator: an integrated system for functional annotation of microRNAs. Nucleic Acids Res. 2008;36:D159–D164. doi: 10.1093/nar/gkm829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maragkakis M, Reczko M, Simossis VA, Alexiou P, Papadopoulos GL, Dalamagas T, Giannopoulos G, Goumas G, Koukis E, Kourtis K, et al. DIANA-microT web server: elucidating microRNA functions through target prediction. Nucleic Acids Res. 2009;37:W273–W276. doi: 10.1093/nar/gkp292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hausser J, Berninger P, Rodak C, Jantscher Y, Wirth S, Zavolan M. MirZ: an integrated microRNA expression atlas and target prediction resource. Nucleic Acids Res. 2009;37:W266–W272. doi: 10.1093/nar/gkp412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nam S, Li M, Choi K, Balch C, Kim S, Nephew KP. MicroRNA and mRNA integrated analysis (MMIA): a web tool for examining biological functions of microRNA expression. Nucleic Acids Res. 2009;37:W356–W362. doi: 10.1093/nar/gkp294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fulci V, Colombo T, Chiaretti S, Messina M, Citarella F, Tavolaro S, Guarini A, Foa R, Macino G. Characterization of B- and T-lineage acute lymphoblastic leukemia by integrated analysis of MicroRNA and mRNA expression profiles. Genes Chromosomes Cancer. 2009;48:1069–1082. doi: 10.1002/gcc.20709. [DOI] [PubMed] [Google Scholar]
- 23.Steuer R, Kurths J, Daub CO, Weise J, Selbig J. The mutual information: detecting and evaluating dependencies between variables. Bioinformatics. 2002;18(Suppl. l2):S231–S240. doi: 10.1093/bioinformatics/18.suppl_2.s231. [DOI] [PubMed] [Google Scholar]
- 24.Kraskov A, Stogbauer H, Grassberger P. Estimating mutual information. Phys. Rev. E Stat. Nonlin. Soft Matter Phys. 2004;69:066138. doi: 10.1103/PhysRevE.69.066138. [DOI] [PubMed] [Google Scholar]
- 25.Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 2004;3:Article 3. doi: 10.2202/1544-6115.1027. [DOI] [PubMed] [Google Scholar]
- 26.Moreau Y, Aerts S, De Moor B, De Strooper B, Dabrowski M. Comparison and meta-analysis of microarray data: from the bench to the computer desk. Trends Genet. 2003;19:570–577. doi: 10.1016/j.tig.2003.08.006. [DOI] [PubMed] [Google Scholar]
- 27.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protocols. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 28.Maglott D, Ostell J, Pruitt KD, Tatusova T. Entrez gene: gene-centered information at NCBI. Nucleic Acids Res. 2005;33:D54–D58. doi: 10.1093/nar/gki031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Parkinson H, Kapushesky M, Kolesnikov N, Rustici G, Shojatalab M, Abeygunawardena N, Berube H, Dylag M, Emam I, Farne A, et al. ArrayExpress update – from an archive of functional genomics experiments to the atlas of gene expression. Nucleic Acids Res. 2009;37:D868–D872. doi: 10.1093/nar/gkn889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jiang Q, Wang Y, Hao Y, Juan L, Teng M, Zhang X, Li M, Wang G, Liu Y. miR2Disease: a manually curated database for microRNA deregulation in human disease. Nucleic Acids Res. 2009;37:D98–D104. doi: 10.1093/nar/gkn714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xiao F, Zuo Z, Cai G, Kang S, Gao X, Li T. miRecords: an integrated resource for microRNA-target interactions. Nucleic Acids Res. 2009;37:D105–D110. doi: 10.1093/nar/gkn851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Plikus MV, Zhang Z, Chuong CM. PubFocus: semantic MEDLINE/PubMed citations analytics through integration of controlled biomedical dictionaries and ranking algorithm. BMC Bioinformatics. 2006;7:424. doi: 10.1186/1471-2105-7-424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rebholz-Schuhmann D, Kirsch H, Arregui M, Gaudan S, Riethoven M, Stoehr P. EBIMed–text crunching to gather facts for proteins from Medline. Bioinformatics. 2007;23:e237–e244. doi: 10.1093/bioinformatics/btl302. [DOI] [PubMed] [Google Scholar]
- 34.Qiang YW, Endo Y, Rubin JS, Rudikoff S. Wnt signaling in B-cell neoplasia. Oncogene. 2003;22:1536–1545. doi: 10.1038/sj.onc.1206239. [DOI] [PubMed] [Google Scholar]
- 35.Peifer M, Polakis P. Wnt signaling in oncogenesis and embryogenesis–a look outside the nucleus. Science. 2000;287:1606–1609. doi: 10.1126/science.287.5458.1606. [DOI] [PubMed] [Google Scholar]
- 36.Bashir T, Pagano M. Aberrant ubiquitin-mediated proteolysis of cell cycle regulatory proteins and oncogenesis. Advances Cancer Res. 2003;88:101–144. doi: 10.1016/s0065-230x(03)88305-7. [DOI] [PubMed] [Google Scholar]
- 37.Chien Y, White MA. RAL GTPases are linchpin modulators of human tumour-cell proliferation and survival. EMBO Rep. 2003;4:800–806. doi: 10.1038/sj.embor.embor899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Garzon R, Pichiorri F, Palumbo T, Visentini M, Aqeilan R, Cimmino A, Wang H, Sun H, Volinia S, Alder H, et al. MicroRNA gene expression during retinoic acid-induced differentiation of human acute promyelocytic leukemia. Oncogene. 2007;26:4148–4157. doi: 10.1038/sj.onc.1210186. [DOI] [PubMed] [Google Scholar]
- 39.Mi S, Lu J, Sun M, Li Z, Zhang H, Neilly MB, Wang Y, Qian Z, Jin J, Zhang Y, et al. MicroRNA expression signatures accurately discriminate acute lymphoblastic leukemia from acute myeloid leukemia. Proc. Natl Acad. Sci. USA. 2007;104:19971–19976. doi: 10.1073/pnas.0709313104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hahne JC, Kummer S, Heukamp LC, Fuchs T, Gun M, Langer B, Von Ruecker A, Wernert N. Regulation of protein tyrosine kinases in tumour cells by the transcription factor Ets-1. Int. J. Oncol. 2009;35:989–996. doi: 10.3892/ijo_00000413. [DOI] [PubMed] [Google Scholar]
- 41.Hrstka R, Coates PJ, Vojtesek B. Polymorphisms in p53 and the p53 pathway: roles in cancer susceptibility and response to treatment. J. Cell. Mol. Med. 2009;13:440–453. doi: 10.1111/j.1582-4934.2008.00634.x. [DOI] [PMC free article] [PubMed] [Google Scholar]