Abstract
ALTER is an open web-based tool to transform between different multiple sequence alignment formats. The originality of ALTER lies in the fact that it focuses on the specifications of mainstream alignment and analysis programs rather than on the conversion among more or less specific formats. In addition, ALTER is capable of identify and remove identical sequences during the transformation process. Besides its user-friendly environment, ALTER allows access to its functionalities in a programmatic way through a Representational State Transfer web service. ALTER’s front-end and its API are freely available at http://sing.ei.uvigo.es/ALTER/ and http://sing.ei.uvigo.es/ALTER/api/, respectively.
INTRODUCTION
Multiple sequence alignments (MSAs) are at the core of many bioinformatic analyses that benefit from the comparison of genomic sequences, from phylogenetic reconstruction to functional prediction (1,2). MSAs can be stored in a large variety of formats (e.g. FASTA, PIR, PHYLIP, NEXUS, etc.), and very often, researchers are obligated to transform between these in order to use different tools. Some conversion utilities have been extremely useful in this regard, the most popular being ReadSeq (http://iubio.bio.indiana.edu/soft/molbio/readseq/java/). Indeed, there are other tools developed mainly for other purposes that can also import and export aligments in several formats, like ReadAl/TrimAl (3), SeaView (4), Se-Al (http://tree.bio.ed.ac.uk/software/seal/) or even ClustalX2 (5), among others. Moreover, projects like BioPython (6) or BioPerl (7) also offer conversion capabilities.
However, the problem with most of these converters is that they—logically—focus on more or less flexible format specifications that are often violated by both developers and users. In fact, during the last years MSA’s formats have ‘evolved’ very much like the sequences they contain, with mutational events consisting of long names, extra spaces, additional carriage returns, etc. Thus, different applications often require or produce particular MSA formats that in fact do not completely fulfill the requirements of the ‘canonical’ formats, often complicating the use of different tools for the analysis of data. For example, ReadSeq and programs like PAML (8) or PAUP* (http://paup.csit.fsu.edu/) fail to read simple alignments produced by ClustalX2 in PHYLIP format. To alleviate these kind of problems, we introduce a web server called ALTER for the program-oriented—rather than format-oriented—conversion between DNA and protein MSA formats. ALTER is free and open to all and there is no login requirement.
FUNCIONALITY
ALTER was designed to accomplish two main objectives: (i) easily convert between MSA formats used by popular tools and (ii) collapse sequences to haplotypes (unique sequences). In order to perform these operations in an intuitive way, ALTER implements a straightforward workflow that easily guides the user through a four-step wizard in which the different options are automatically activated when the required information is available. In addition, ALTER provides an easy-to-follow on-line help as well as many sample MSA data for testing purposes.
Program workflow
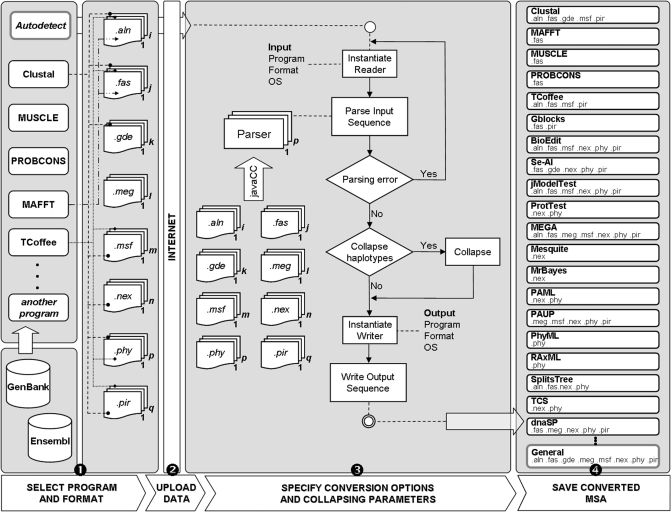
The use of ALTER typically implies four simple steps: (i) format/program identification, (ii) data load, (iii) definition of conversion parameters and (iv) storage of the generated file (Figure 1).
Figure 1.
Schematic ALTER workflow. The user can select between different input alignment programs and formats, and obtain a MSA specifically formatted for a particular program.
The process of converting a given MSA in ALTER starts with the selection of the source program and/or the current format. If the user is not confident about this information, the server can try to auto detect the format of the input file.
Next, the user has to specify the operating system (OS) under which the input file was generated and upload it, or alternatively directly paste the data. In order to process the input MSA, ALTER first instantiates an appropriate sequence reader for both the input format and program. For each program/format pair, there is a specific parser generated from a formal grammar via JavaCC technology. Regardless of the possibility to reuse grammars among programs that utilize the same format, ALTER has been designed to be able to associate a different grammar for each program/format pair in order to tackle potential differences. If the user has selected the ‘auto detect’ option, a program-independent grammar is used instead. If there are syntax errors on the input sequences, the parser reports precise information about them and the process aborts.
Once the input MSA has been successfully read, ALTER can perform an optional step to identify redundant sequences and collapse them into haplotypes. Finally, an appropriate writer for the output program/format/OS is instantiated in order to generate the converted MSA, taking into account different parameters. These allow the user to (i) generate sequential or interleaved sequences (in NEXUS and PHYLIP formats), (ii) use lower case for residues, (iii) use match characters (‘.’) to indicate that the same residue is located at the same position of the first sequence and (iii) generate the sum of the number of residues at each sequence line (ALN format). In addition, the collapsing step can be configured to (i) treat gaps as missing data, (ii) consider missing data as differences between sequences and (iii) define a maximum limit of differences to collapse sequences. It is also possible to generate a program-independent conversion using only the canonical format specification.
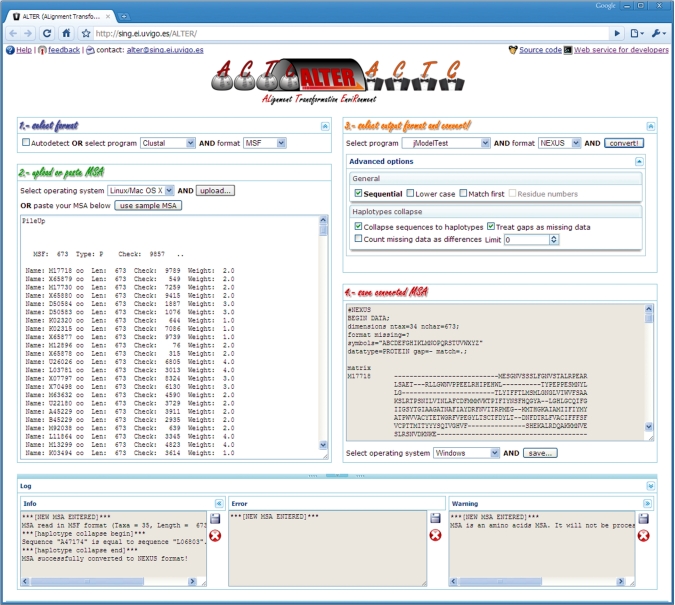
Every time a new conversion job finished without errors, the output file is displayed and a download button is activated. All the relevant information related to the process of loading and recognizing the input MSA is automatically categorized (info, error, warning) and displayed to the final user by using informative log panels (Figure 2).
Figure 2.
Example of a MSA conversion in ALTER. The ‘Info panel’ in the log area shows information related with the process carried out. Help, support for feedback and contact information options are available from the upper left area. Source code and a description of web services are available from the upper right area.
Supported MSA formats/programs
ALTER supports a variety of specific MSA formats provided by popular alignment tools and accepted by a variety of analysis programs. Currently, the focus is on molecular evolution, but different tools can be easily added on request. The list of programs supported include alignment, alignment filtering, sequence edition, model selection, phylogenetic, network and population genetics software (Table 1).
Table 1.
List programs/formats supported by ALTER
| Tools | Supported formats |
|---|---|
| INPUT: multiple sequence alignment programs | |
| Clustal (10) | ALN, FASTA, GDE, MSF, NEXUS, PHYLIP, PIR |
| MAFFT (11) | ALN, FASTA |
| TCoffee (12) | ALN, FASTA, MSF, PHYLIP, PIR |
| MUSCLE (13) | ALN, FASTA, MSF, PHYLIP |
| PROBCONS (14) | ALN, FASTA |
| OUTPUT: alignment | |
| Clustal | ALN, FASTA, GDE, MSF, PIR |
| MAFFT | FASTA |
| MUSCLE | FASTA |
| PROBCONS | FASTA |
| TCoffee | ALN, FASTA, MSF, PIR |
| OUTPUT: alignment filtering | |
| Gblocks (15) | FASTA, PIR |
| OUTPUT: sequence edition | |
| BioEdit (16) | ALN, FASTA, MSF, NEXUS, PHYLIP, PIR |
| Se-Ala | FASTA, GDE, NEXUS, PHYLIP, PIR |
| OUTPUT: model selection | |
| jModelTest (17) | ALN, FASTA, MSF, NEXUS, PHYLIP, PIR |
| ProtTest (18) | NEXUS, PHYLIP |
| OUTPUT: phylogenetic analysis | |
| MEGA (19) | ALN, FASTA, MEGA, MSF, NEXUS, PHYLIP, PIR |
| Mesquiteb | NEXUS |
| MrBayes (20) | NEXUS |
| PAML (8) | NEXUS, PHYLIP |
| PAUP (21) | MEGA, MSF, NEXUS, PHYLIP, PIR |
| PhyML (22) | PHYLIP |
| RaxML (23) | PHYLIP |
| OUTPUT: phylogenetic networks | |
| SplitsTree (24) | ALN, FASTA, NEXUS, PHYLIP |
| TCS (25) | NEXUS, PHYLIP |
| OUTPUT: population genetics | |
| DnaSP (26) | FASTA, MEGA, NEXUS, PHYLIP, PIR |
| OUTPUT: General | |
| standard specification | ALN, FASTA, GDE, MEGA, MSF, NEXUS, PHYLIP, PIR |
Web services
In addition to the functionality provided by the end user front-end, ALTER also implements a web service that allows developers to transform multiple alignment sequences directly in ALTER within their own algorithms and programs (http://sing.ei.uvigo.es/ALTER/api/). Essentially, ALTER’s API offers a unique convert function with multiple parameters plus some metadata functions giving information about the formats and options currently supported. Table 2 summarizes the API functionality.
Table 2.
Core functionality provided by ALTER’s RESTful API
| Function | Description |
|---|---|
| Convert | Converts an input sequence from one format to another. This function is accessed via HTTP POST where both the sequence and parameters should be sent to the server. |
| Metadata functions | |
| List OSs | Lists the available OSs to read files from. |
| URL: http://sing.ei.uvigo.es/ALTER/api/so | |
| List input programs | Lists the currently supported input programs. |
| URL: http://sing.ei.uvigo.es/ALTER/api/input/programs | |
| List input formats | Lists the currently supported input formats. |
| URL: http://sing.ei.uvigo.es/ALTER/api/input/formats | |
| List output programs | Lists the currently supported output programs. |
| URL: http://sing.ei.uvigo.es/ALTER/api/output/programs | |
| List output formats | Lists the currently supported output formats. |
| URL: http://sing.ei.uvigo.es/ALTER/api/output/formats | |
| List output formats for a specific program | Lists the supported output formats for a given output program. |
| Example URL: http://sing.ei.uvigo.es/ALTER/api/output/paml/formats | |
| List options for output program and format | Lists the supported options for a given output program and format. |
| Example URL: http://sing.ei.uvigo.es/ALTER/api/output/paml/nexus/options | |
Supported platforms
ALTER runs on a standard Tomcat 5.5 Web application server. Currently, ALTER has been successfully tested in Internet Explorer 7, Firefox 3, Opera 9.62 and Safari 3 browsers working on Windows XP/Vista, Ubuntu Linux 8.04 version and Mac OSX 10.5 of Intel architecture.
IMPLEMENTATION
ALTER is implemented as an AJAX-enabled web application programmed in the J2SE 1.5 Java language. The ZK development framework (http://www.zkoss.org) was used to construct the user interface and to give support to JavaCC for parsing input MSA. JavaCC is a parser and a lexical analyzer generator, that is, it reads a formal description of a language (grammar) and generates code to parse instances of it. It can be see as the Java counterpart of the Lex/Flex and Yacc/Bison tools. Using JavaCC it is possible to (i) isolate the specific sequence format description in independent grammar files and (ii) generate precise error messages during parsing (9).
ALTER also implements a REST-based programming interface. Like any RESTful web service, operations are performed via web queries with a well-defined URL structure. Currently, the server gives access to the main sequence conversion functionality as well as to a set of reflective functions intended to get updated information about the supported programs and formats. This server module was implemented following the JAX-RS 1.0 (Java API for RESTful Web Services) by using the implementation found in the Apache CXF library.
CONCLUSIONS
Current MSA conversion tools understandably focus on the translation among ‘canonical’ formats, but in many instances are not of much help for users, which are interested in working with particular programs that use idiosyncratic format variations. In order to alleviate this drawback, we introduce a web server called ALTER for the program-oriented—rather than format-oriented—conversion between different DNA and protein MSA formats. In addition, ALTER is able to ‘collapse’ sequences to haplotypes—unique sequences—indicating which sequence corresponds to which haplotype. Eliminating this redundancy can be very helpful, for example, to speed up phylogenetic analyses.
FUNDING
European Research Council (ERC-2007-Stg 203161-PHYGENOM to D.P.); Spanish Ministry of Science and Education (BFU2009-08611 to D.P.); Xunta de Galicia (PGIDIT07PXIB310202PR to D.P.); INBIOMED initiative, Angeles Alvariño fellowship (to D.G-P.); University of Vigo (09VIB10 to F.F-.R.). Funding for open access charge: European Research Council (ERC-2007-Stg 203161-PHYGENOM to D.P.).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors want to thank all the beta testers, especially those from the Bioinformatics and Molecular Evolution group at the University of Vigo.
REFERENCES
- 1.Posada D, editor. Bioinformatics for DNA sequence analysis. New York, NY, USA: Humana Press; 2009. [DOI] [PubMed] [Google Scholar]
- 2.Kemena C, Notredame C. Upcoming challenges for multiple sequence alignment methods in the high-throughput era. Bioinformatics. 2009;25:2455–2465. doi: 10.1093/bioinformatics/btp452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Capella-Gutierrez S, Silla-Martinez JM, Gabaldon T. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics. 2009;25:1972–1973. doi: 10.1093/bioinformatics/btp348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gouy M, Guindon S, Gascuel O. SeaView version 4: a multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol. Biol. Evol. 2010;27:221–224. doi: 10.1093/molbev/msp259. [DOI] [PubMed] [Google Scholar]
- 5.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 6.Cock PJ, Antao T, Chang JT, Chapman BA, Cox CJ, Dalke A, Friedberg I, Hamelryck T, Kauff F, Wilczynski B, et al. Biopython: freely available Python tools for computational molecular biology and bioinformatics. Bioinformatics. 2009;25:1422–1423. doi: 10.1093/bioinformatics/btp163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stajich JE, Block D, Boulez K, Brenner SE, Chervitz SA, Dagdigian C, Fuellen G, Gilbert JG, Korf I, Lapp H, et al. The Bioperl toolkit: perl modules for the life sciences. Genome Res. 2002;12:1611–1618. doi: 10.1101/gr.361602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang Z. PAML 4: phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 2007;24:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- 9.Metsker SJ. Building Parsers With Java. Boston, MA, USA: Addison-Wesley Professional; 2001. [Google Scholar]
- 10.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Katoh K, Kuma K, Toh H, Miyata T. MAFFT version 5: improvement in accuracy of multiple sequence alignment. Nucleic Acids Res. 2005;33:511–518. doi: 10.1093/nar/gki198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Notredame C, Higgins DG, Heringa J. T-Coffee: a novel method for fast and accurate multiple sequence alignment. J. Mol. Biol. 2000;302:205–217. doi: 10.1006/jmbi.2000.4042. [DOI] [PubMed] [Google Scholar]
- 13.Edgar RC. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics. 2004;5:113. doi: 10.1186/1471-2105-5-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Do CB, Mahabhashyam MS, Brudno M, Batzoglou S. ProbCons: probabilistic consistency-based multiple sequence alignment. Genome Res. 2005;15:330–340. doi: 10.1101/gr.2821705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Castresana J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol. Biol. Evol. 2000;17:540–552. doi: 10.1093/oxfordjournals.molbev.a026334. [DOI] [PubMed] [Google Scholar]
- 16.Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999;41:95–98. [Google Scholar]
- 17.Posada D. jModelTest: phylogenetic model averaging. Mol. Biol. Evol. 2008;25:1253–1256. doi: 10.1093/molbev/msn083. [DOI] [PubMed] [Google Scholar]
- 18.Abascal F, Zardoya R, Posada D. ProtTest: selection of best-fit models of protein evolution. Bioinformatics. 2005;21:2104–2105. doi: 10.1093/bioinformatics/bti263. [DOI] [PubMed] [Google Scholar]
- 19.Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- 20.Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- 21.Swofford DL. PAUP*: Phylogenetic analysis using parsimony (*and Other Methods) 2000 Sunderland, MA, USA. [Google Scholar]
- 22.Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- 23.Stamatakis A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22:2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- 24.Huson DH. SplitsTree: analyzing and visualizing evolutionary data. Bioinformatics. 1998;14:68–73. doi: 10.1093/bioinformatics/14.1.68. [DOI] [PubMed] [Google Scholar]
- 25.Clement M, Posada D, Crandall KA. TCS: a computer program to estimate gene genealogies. Mol. Ecol. 2000;9:1657–1659. doi: 10.1046/j.1365-294x.2000.01020.x. [DOI] [PubMed] [Google Scholar]
- 26.Librado P, Rozas J. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 2009;25:1451–1452. doi: 10.1093/bioinformatics/btp187. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.