Abstract
miRNAs are small, non-coding RNA that negatively regulate gene expression at post-transcriptional level, which play crucial roles in various physiological and pathological processes, such as development and tumorigenesis. Although deep sequencing technologies have been applied to investigate various small RNA transcriptomes, their computational methods are far away from maturation as compared to microarray-based approaches. In this study, a comprehensive web server mirTools was developed to allow researchers to comprehensively characterize small RNA transcriptome. With the aid of mirTools, users can: (i) filter low-quality reads and 3/5′ adapters from raw sequenced data; (ii) align large-scale short reads to the reference genome and explore their length distribution; (iii) classify small RNA candidates into known categories, such as known miRNAs, non-coding RNA, genomic repeats and coding sequences; (iv) provide detailed annotation information for known miRNAs, such as miRNA/miRNA*, absolute/relative reads count and the most abundant tag; (v) predict novel miRNAs that have not been characterized before; and (vi) identify differentially expressed miRNAs between samples based on two different counting strategies: total read tag counts and the most abundant tag counts. We believe that the integration of multiple computational approaches in mirTools will greatly facilitate current microRNA researches in multiple ways. mirTools can be accessed at http://centre.bioinformatics.zj.cn/mirtools/ and http://59.79.168.90/mirtools.
INTRODUCTION
MicroRNAs (miRNAs) are endogenous, non-coding RNAs of ∼22 nt in length that regulate gene expression post-transcriptionally by directly guiding RNA-induced silencing complex (RISC) to cognate mRNA targets (1,2). Particularly, the complementary sequence in the ‘seed region’ (6–8 bp) at the 5′-end of the miRNA–mRNA heteroduplex provide an obvious clue that there are specified interactions between miRNA and its targets. Recent studies have documented the potent pro- and anti-tumorigenic activities of specific miRNAs both in vitro and in vivo (3). Potential influences have been verified in more widespread biological processes, such as viral infections, cardiovascular diseases and neurological and muscular disorders, as well as tumorigenesis (4,5). Consequently, identifying comprehensive sets and differentially expressed miRNAs across tissues and cell lines is attracting considerably more attention.
The emergence of high-throughput next-generation sequencing technologies has dramatically changed the speed of all aspects of sequencing in a rapid and cost-effective fashion, which can permit unbiased, quantitive and in-depth investigation of the small RNA transcriptome that has been previously possible (6,7). Compared to previous hybridization-based methods, deep sequencing approaches have several advantages, such as high resolution, high yield and reduced complexity of experimental procedures. However, deep sequencing-based expression analysis is still in its infancy and has substantial informatics challenges for lack of efficient and flexible tools to handle and analyze a huge scale of short sequences (8,9).
Till now, there are several public tools that are available for miRNA transcriptomic analysis from deep sequencing data. The miRDeep package was developed to discover active known or novel miRNAs from deep sequencing data, and it includes scripts to preprocess raw reads and also algorithms to analyze and score miRNA expression data (10). miRExpress is a stand-alone software package implemented for generating miRNA expression profiles from high-throughput sequencing of microRNAs (11). SeqBuster is a web-based tool to allow users to investigate the miRNA variants or isomers hidden in large-scale small RNA datasets (12). miRanalyzer is a web server tool that can detect all known miRNA sequences annotated in miRBase and predict novel miRNAs based on a machine-learning approach (13). UEA small RNA tools were designed to the analysis of high-throughput small RNA data, such as identification of miRNAs and their targets, and comparison expression levels in specific small RNA loci (14). deepBase is a comprehensive database developed to annotate and discover small RNAs from transcriptomic data (15). However, to the best of our best knowledge, none of the currently available tools provides web-based approaches to analyze multiple transcriptomes. In this study, we present a novel web server mirTools, which can (i) filter low-quality reads and adapters; (ii) classify large-scale short reads into known categories; (iii) predict novel miRNAs and their secondary structures; and (iv) identify significantly differentially expressed miRNAs. We believe that the integration of multiple computational approaches in mirTools will greatly facilitate current microRNA researches.
mirTools ANALYSIS WORKFLOW
Read filter
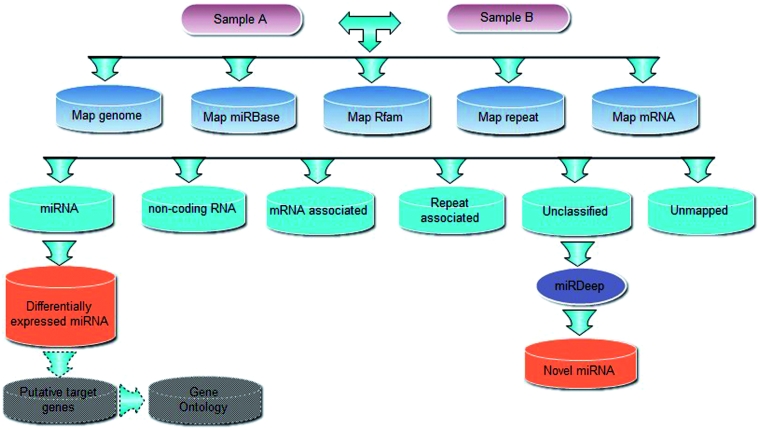
The current procedure of mirTools used to annotate small RNA transcriptome by high-throughput sequencing is shown in Figure 1. Briefly, for deep sequencing reads produced by Illumina Genome Analyzer or 454 FLX instrument, low-quality reads must be filtered out to exclude those most likely to represent sequencing errors and 3/5′ adaptor sequences. Subsequently, they are trimmed into clean full-length reads and formatted into a non-redundant FASTA file. The occurrence of each unique sequence read is counted as sequence tag and the number of reads for each tag reflects its relative expression level.
Figure 1.
The small RNA annotation workflow of mirTools.
Small RNA annotation
All unique sequence tags that pass through above filtering criteria are mapped onto the reference genome using the SOAP program (16). Subsequently, these unique sequence tags are also aligned against miRBase (17), Rfam (18), repeat database produced by RepeatMasker (19) and the coding genes of the reference genome. In this way, the unique sequence tags can be classified into the following categories: known miRNA, degradation fragments of non-coding RNA, genomic repeats and mRNA. In case of conflict, a hierarchy is conducted to assign the tag into a unique category, which starts with non-coding RNA, then known miRNA and followed by repeat associated RNA and mRNA. Sequences that are assigned to none of these annotations but can be mapped to the reference genome are classified as ‘unclassified’.
Differential expression detection
To compare differentially expressed miRNAs between multiple samples, read count of each identified miRNA is normalized to the total number of miRNA read counts that are matched to the reference genome in each sample. The statistical significance (P-value) is inferred based on a Bayesian method (20), which was developed for analyzing digital gene expression profiles and could account for the sampling variability of tags with low counts. In default, a specific miRNA will be deemed to be significantly differentially expressed when the P-value given by this method is ≤0.01 and there is at least a 2-fold change in normalized sequence counts.
Novel miRNA prediction
Sequences that do not fall into above annotation categories but matched on the reference genome are used to detect candidate novel miRNA genes. In default, 100 nucleotides of genomic sequence flanking each side of these sequences are extracted and their RNA secondary structures are predicted using RNAfold (21). Novel miRNAs are identified by folding the flanking genomic sequence using the miRDeep program.
IMPLEMENTATION
The web design scheme of mirTools follows our previous integrated PGA4genomics pipeline (22), which was constructed based on open source softwares. Briefly, mirTools is programmed in Perl and the web server is hosted on an Apache 2.0 HTTP server under a Linux operating system. The front-end, implemented in PHP language scripts, provides results upon request for users once a job is finished. The server is equipped with four Quad-Core AMD processors (2.2 GHz each) and 32 GB of RAM. Meanwhile, mirTools has a queuing module to control user-submitted jobs, which only executes two jobs in parallel and extra ones will be put into a queue. The web application is implemented in an operating-system independent way and has been successfully tested in Microsoft Internet Explorer 8.0 and Firefox 2/3 (under different versions of Linux, Microsoft Windows and MacOS).
DATA INPUT
mirTools provides a simple and user-friendly interface to allow users to extensively annotate the small RNA transcriptome generated from high-throughput sequencing (Figure 1). The input requirement of mirTools is a trimmed FASTA file obtained from pre-processed raw data as follows:
>UniTag-009_×80
CATTTATTATTTATCTTATTCCTTCTTCTTTTTTA
Where, ‘UniTag-009’ represents a user-definable unique ID for reads with identical sequences. The ‘×80’ indicates that this tag (UniTag-009) has occurred 80 times in the sequenced sample. Both of them must be linked with an underline. In such a way, a gigabyte-scale file with unprecedented amounts of reads can be significantly reduced to an acceptable size for a web server tool. To generate the desired input format, we provide a command-line Perl script (http://centre.bioinformatics.zj.cn/mirtools/adaptortrim.php), through which low quality reads, 3/5′ adaptor sequences and polyA can be easily filtered from raw sequenced data.
mirTools server runs in a two-mode architecture on a centralized platform according to the number of uploaded samples: single and multiple. In the ‘single’ mode, the server will analyze and annotate small RNAs embed in the sample. In the ‘multiple’ mode, additionally, the server identifies differential expression miRNAs between pair designed samples. It should be noted that, currently, the maximum allowable size of input file uploaded is limited to 10 MB, which can be in FASTA format or compressed in zip or gz format containing the FASTA file. In addition, to satisfy personalized small RNA annotation, a number of important parameters are provided in both modes. Users can set a length interval in advance and only the tag sequences within this length interval will be considered for downstream analysis. mirTools allow users to define the number of allowed mismatches (at most two mismatches) between the tag sequence and genomic sequences, as well as other annotation information. When predicting novel miRNAs, the flanking sequence length of the query is supported. To detect differentially expressed miRNAs between samples, the desired statistical significance of interest with P-value threshold and fold change in normalized sequence counts can be defined by users. A specific miRNA will be deemed to be significantly differentially expressed when both the P-value and the fold change are satisfied.
After data submission, a typical run may take 1–2 h to finish, depending on the data size, reference genome size and queuing jobs. If users provide a valid email address, mirTools will send out a notification with a URL of the data archive when the job is completed. In addition, users can retrieve their results from the stored jobs (no longer than one month) with a unique ID randomly generated by the server for each job.
DATA OUTPUT
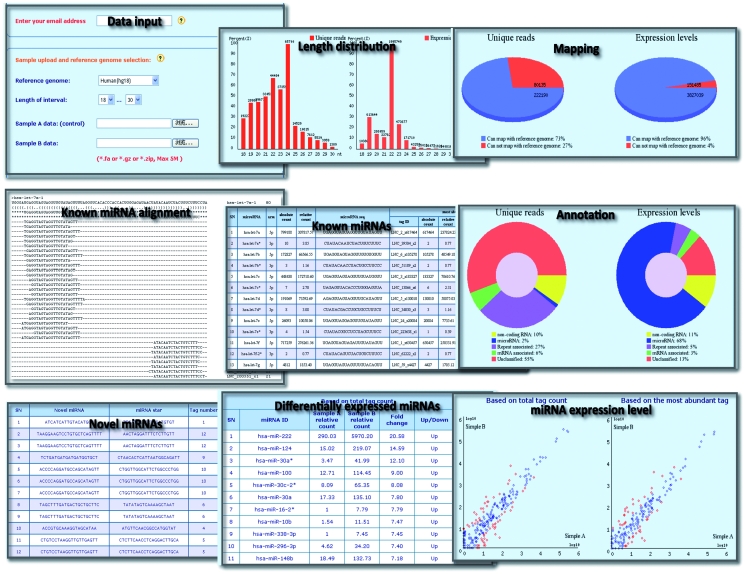
A typical output of mirTools consists of six parts: length distribution, reference genome mapping, annotation, known miRNAs, novel miRNAs and differentially expressed miRNAs (Figure 2). All these components are well organized with examples to facilitate users with the correct input and expected results.
Figure 2.
Screenshots of the mirTools web interface.
The first two parts give an overview of the length distribution of miRNAs and their mapping ratios against the reference genome. mirTools plots both the unique read distribution and expression levels (the number of reads for each tag reflects its relative abundance). This is useful to allow users to easily determine the efficiency of the deep sequencing procedure for miRNA detection and to simultaneously compare length distributions between samples.
The third part summarizes the percentage of small RNAs classified into different functional categories. Currently, mirTools assign the small RNA sequences into one of following categories: known miRNA, degradation fragments of non-coding RNA (tRNA, rRNA, snRNA/snoRNA, etc.), genomic repeat, mRNA and unclassified. In the fourth part, mirTools provides a detailed annotation for each known miRNA. In the left, a table shows known miRNA ID, 5′/3′ arm, absolute count, relative counts (normalized to the total number of miRNA reads and then multiplied by 106), miRNA sequence and most abundant tags with Tag ID, absolute/relative counts and corresponding tag sequence. Visual sequence alignments matched to a specific miRNA are listed in tabulated text files in the right.
In the fifth part, novel miRNAs identified by miRDeep are provided, which contain novel miRNA sequence, tag number, tag count and responding hairpin structure, which is displayed in a SVG format and thus requires a SVG plug-in to be installed in client’s computers. In the last part, the relative expression level of all miRNAs is illustrated in a scatter plot with red dots representing differentially expressed miRNAs. Meanwhile, detailed annotation of these differentially expressed miRNAs is provided, including miRNA ID, relative sequenced count, fold change, up-regulated/down-regulated and P-value. It should be noted that mirTools employs two different measures to evaluate miRNA expression levels: one is based on the total tag count (#specific miRNA tags/#total miRNA sequence tags) and the other is based on the most abundant tag count (#the most abundant tag of specific miRNA /#total miRNA sequence tags).
CASE STUDIES
To evaluate the performance of mirTools, the small RNA transcriptomes from the human embryonic stem cell hESC and EB libraries were downloaded from ftp://ftp03.bcgsc.ca/public/hESC_miRNA (23). mirTools identified 533 and 573 known miRNAs from hESC and EB, respectively (http://centre.bioinformatics.zj.cn/mirtools/download2.php?id=1264223745), which are a little more than the numbers identified in previous study. This is due to recent updates in miRBase (version 14) used in mirTools. When detecting differentially expressed miRNAs, we set the same parameters (P < 0.001, fold change ≥ 1.5) as Morin’s study. As shown in Supplementary Figure S1A, the expression level changes identified by both studies are highly correlated (Pearson’s correlation coefficient=0.96).
Additionally, two small RNA transcriptome data sets derived from our lab were performed. This experiment was designed to characterize the miRNA expression profiles underlying the progression of androgen-dependent (LNCaP) to androgen-independent prostate cancer (LNCaP-AI). Standard protocols were used for small RNA preparation and Illumina sequencing. In total, 9 107 833 and 10 083 251 raw reads were generated for LNCaP and LNCaP-AI, respectively (data not shown). Following the mirTools’s workflow, we filtered low-quality reads and adapters, and mapped them onto the human reference genome (hg18), and annotated them using currently available miRNA databases. Additionally, a detailed list of significantly differentially expressed microRNAs was identified. The summary output of mirTools run on these data was shown in the web server (http://centre.bioinformatics.zj.cn/mirtools/download2.php?id=1259926142). To further validate these differentially expressed miRNAs, 29 of them were selected to perform qRT–PCR analysis. As shown in Supplementary Figure S1B, a strong correlation (Pearson’s correlation coefficient=0.91) was observed between the Illumina deep sequencing data and the qRT–PCR measurements, indicating the robustness of deep sequencing-based expression analysis obtained from mirTools.
PERSPECTIVES
In transcriptomic studies, high-throughput sequencing technologies generate an overwhelming amount of raw reads, which present substantial informatics challenges for a lack of efficient and flexible tools. In order to address these challenges, mirTools was developed toward a fully automated and easy to use web service suitable for small RNA transcriptome analyses. Several steps in mirTools are computationally intensive, which make us restrict upload data size to avoid overloading the source website. In the future, we will improve the server’s computational efficiency and decrease the input file size limit. mirTools depend on a precomputed annotation of reference genome. Currently, it supports 15 most commonly studied model organisms, ranging from vertebrates, invertebrates, to plants. More reference genomes will be integrated in future update. In addition, mirTools will intend to implement miRanda (http://www.microrna.org/)and RNAhybrid (http://bibiserv.techfak.uni-bielefeld.de/rnahybrid/) to predict the targeted genes of identified miRNAs. Meanwhile, Gene Ontology terms and KEGG pathways of targeted genes will also be integrated to evaluate target gene functions.
It is believed that with simplicity, robustness and accessibility of mirTools can serve as a useful tool for comprehensive characterization of the small RNA transcriptomes obtained from high-throughput sequencing. In general, mirTools can fill the gaps between high-throughput sequencing and extensive bioinformatics analysis, and alleviate the limitation in miRNA profiling and discovery.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Funding for open access charge: National High Technology Research and Development Program of China (2006AA02A304); Major State Basic Research Development Program of China (2007CB512302).
Conflict of interest statement. None declared.
Supplementary Material
REFERENCES
- 1.Kim VN, Nam JW. Genomics of microRNA. Trends Genet. 2006;22:165–173. doi: 10.1016/j.tig.2006.01.003. [DOI] [PubMed] [Google Scholar]
- 2.Brennecke J, Stark A, Russell RB, Cohen SM. Principles of microRNA-target recognition. PLoS Biol. 2005;3:e85. doi: 10.1371/journal.pbio.0030085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat. Rev. Cancer. 2006;6:857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 4.Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 5.He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nat. Rev. Genet. 2004;5:522–531. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- 6.Creighton CJ, Reid JG, Gunaratne PH. Expression profiling of microRNAs by deep sequencing. Brief Bioinform. 2009;10:490–497. doi: 10.1093/bib/bbp019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fahlgren N, Sullivan CM, Kasschau KD, Chapman EJ, Cumbie JS, Montgomery TA, Gilbert SD, Dasenko M, Backman TW, Givan SA, et al. Computational and analytical framework for small RNA profiling by high-throughput sequencing. RNA. 2009;15:992–1002. doi: 10.1261/rna.1473809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McPherson JD. Next-generation gap. Nat. Methods. 2009;6:S2–S5. doi: 10.1038/nmeth.f.268. [DOI] [PubMed] [Google Scholar]
- 9.Horner DS, Pavesi G, Castrignano T, De Meo PD, Liuni S, Sammeth M, Picardi E, Pesole G. Bioinformatics approaches for genomics and post genomics applications of next-generation sequencing. Brief. Bioinform. 2010;11:181–197. doi: 10.1093/bib/bbp046. [DOI] [PubMed] [Google Scholar]
- 10.Friedlander MR, Chen W, Adamidi C, Maaskola J, Einspanier R, Knespel S, Rajewsky N. Discovering microRNAs from deep sequencing data using miRDeep. Nat. Biotechnol. 2008;26:407–415. doi: 10.1038/nbt1394. [DOI] [PubMed] [Google Scholar]
- 11.Wang WC, Lin FM, Chang WC, Lin KY, Huang HD, Lin NS. miRExpress: analyzing high-throughput sequencing data for profiling microRNA expression. BMC Bioinformatics. 2009;10:328. doi: 10.1186/1471-2105-10-328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pantano L, Estivill X, Marti E. SeqBuster, a bioinformatic tool for the processing and analysis of small RNAs datasets, reveals ubiquitous miRNA modifications in human embryonic cells. Nucleic Acids Res. 2010;38:e34. doi: 10.1093/nar/gkp1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hackenberg M, Sturm M, Langenberger D, Falcon-Perez JM, Aransay AM. miRanalyzer: a microRNA detection and analysis tool for next-generation sequencing experiments. Nucleic Acids Res. 2009;37:W68–W76. doi: 10.1093/nar/gkp347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Moxon S, Schwach F, Dalmay T, Maclean D, Studholme DJ, Moulton V. A toolkit for analysing large-scale plant small RNA datasets. Bioinformatics. 2008;24:2252–2253. doi: 10.1093/bioinformatics/btn428. [DOI] [PubMed] [Google Scholar]
- 15.Yang JH, Shao P, Zhou H, Chen YQ, Qu LH. deepBase: a database for deeply annotating and mining deep sequencing data. Nucleic Acids Res. 2009;38:D123–D130. doi: 10.1093/nar/gkp943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li R, Yu C, Li Y, Lam TW, Yiu SM, Kristiansen K, Wang J. SOAP2: an improved ultrafast tool for short read alignment. Bioinformatics. 2009;25:1966–1967. doi: 10.1093/bioinformatics/btp336. [DOI] [PubMed] [Google Scholar]
- 17.Griffiths-Jones S, Saini HK, van Dongen S, Enright AJ. miRBase: tools for microRNA genomics. Nucleic Acids Res. 2008;36:D154–D158. doi: 10.1093/nar/gkm952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Griffiths-Jones S, Moxon S, Marshall M, Khanna A, Eddy SR, Bateman A. Rfam: annotating non-coding RNAs in complete genomes. Nucleic Acids Res. 2005;33:D121–D124. doi: 10.1093/nar/gki081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tarailo-Graovac M, Chen N. Using RepeatMasker to identify repetitive elements in genomic sequences. Curr. Protoc. Bioinform. 2009 doi: 10.1002/0471250953.bi0410s25. Chapter 4, Unit 4. 108. [DOI] [PubMed] [Google Scholar]
- 20.Audic S, Claverie JM. The significance of digital gene expression profiles. Genome Res. 1997;7:986–995. doi: 10.1101/gr.7.10.986. [DOI] [PubMed] [Google Scholar]
- 21.Hofacker IL. Vienna RNA secondary structure server. Nucleic Acids Res. 2003;31:3429–3431. doi: 10.1093/nar/gkg599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhao F, Hou H, Bao Q, Wu J. PGA4genomics for comparative genome assembly based on genetic algorithm optimization. Genomics. 2009;94:284–286. doi: 10.1016/j.ygeno.2009.06.006. [DOI] [PubMed] [Google Scholar]
- 23.Morin RD, O'Connor MD, Griffith M, Kuchenbauer F, Delaney A, Prabhu AL, Zhao Y, McDonald H, Zeng T, Hirst M, et al. Application of massively parallel sequencing to microRNA profiling and discovery in human embryonic stem cells. Genome Res. 2008;18:610–621. doi: 10.1101/gr.7179508. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.