Abstract
Alternative splicing (AS) is a post-transcriptional process considered to be responsible for the huge diversity of proteins in higher eukaryotes. AS events are regulated by different splicing factors (SFs) that bind to sequence elements on the RNA. SFmap is a web server for predicting putative SF binding sites in genomic data (http://sfmap.technion.ac.il). SFmap implements the COS(WR) algorithm, which computes similarity scores for a given regulatory motif based on information derived from its sequence environment and its evolutionary conservation. Input for SFmap is a human genomic sequence or a list of sequences in FASTA format that can either be uploaded from a file or pasted into a window. SFmap searches within a given sequence for significant hits of binding motifs that are either stored in our database or defined by the user. SFmap results are provided both as a text file and as a graphical web interface.
INTRODUCTION
Alternative splicing (AS) is a post-transcriptional process that generates multiple mRNA isoforms from the same gene. In recent years, AS has emerged as one of the major sources of diversity and functional complexity in humans and other vertebrates (1). Misregulation of AS is also largely associated with human diseases (2). The AS process requires a core splicing mechanism that binds to splicing signals around the exon/intron junctions. Core splicing signals include: (i) the canonical 5′-splice site (‘GU’ in humans) and the 3′-splice site (‘AG’ in humans), located at the 5′- and 3′-ends of the intron, respectively; (ii) the polypyrimidine tract (PPT) located upstream of the 3′-splice site; and (iii) the branch site (conserved ‘A’ in humans) located upstream of the PPT. A web server for mapping splicing signals is available at http://sroogle.tau.ac.il (3). The core splicing signals, however, are not sufficient for accurate recognition of exon/intron junctions. It is well established that additional cis-regulatory elements exist in the exons and the flanking introns that bind to different RNA-binding proteins, acting as splicing factors (SFs) (4). In general, the cis-regulatory elements are relatively short and degenerative sequences, and in many cases, are present in multiple copies on the RNA (5). These SFs can act as positive or negative effectors of the splicing reaction by interacting differentially with exonic or intronic splicing enhancers (ESEs/ISEs) and silencers (ESSs/ISSs), respectively.
In recent years, several methodologies for identifying SF binding sites have been developed (6–10); for review see (11). Most of the available methods for detecting cis-regulatory splicing elements, such as RESCUE-ESE (relative enhancer and silencer classification by unanimous enrichment) (7), PESE (8) and the method by Goren et al. (10), use a de novo prediction approach for detecting motifs enriched in splicing regulatory regions. These methods are generally designed to detect cis-regulatory elements; however, they do not attempt to assign a specific trans-factor to a detected cis-regulatory motif. For example, the RESCUE-ESE web server http://genes.mit.edu/burgelab/rescue-ese/ searches for hexamers that are associated with ESE based on their higher frequency of occurrence in exons versus introns, as well as their significantly higher frequency in exons with weak (non-consensus) splice sites compared to exons with strong (consensus) splice sites (7). A different approach is applied using ESEfinder http://rulai.cshl.edu (6). The ESEfinder web site presents a search interface for detecting significant matches of five ESEs related to four SR proteins: SF2/ASF, SC35, SRp40 and SRp55. The ESEs position specific scoring matrices were derived using functional systematic evolution of ligands by exponential enrichment (12). Another strategy RegRNA http://regrna.mbc.nctu.edu.tw/ was recently developed for identifying different RNA regulatory motifs based on homology to known motifs(13).
Recently, we developed a novel computational approach for predicting and mapping SF binding sites of known SFs that considers both the genomic environment and the evolutionary conservation of SF cis-regulatory elements. The method was used to test experimentally validated sequences showing a high accuracy of 93% and a relatively low false-positive rate of 1% on the tested data. In addition, the method was applied to different sets of exons and introns, and detected an enrichment of SF binding sites in different types of AS events, such as cassette exons, alternative 5′-splice sites and alternative 3′-splice sites (14). Here, we present a web service for mapping SF binding sites on human genomic sequences. As a default, SFmap searches for experimentally defined binding motifs (stored in our database) within the given sequence. In addition to the motifs available on the web server, the user may provide any other RNA motif of interest (of length 4–10 nt). SFmap output includes a link to the results in two different formats, both in text and in a visualized display of the motifs as custom annotation tracks in the UCSC human genome (HG) browser (15). The SFmap web service is free and open to all users with no login requirement.
SFmap METHODOLOGY
Accurate mapping of SF binding sites is challenging, mainly due to the very low information content present in these relatively short and degenerative sequences (4). To enable accurate prediction of SF binding sites, the SFmap server implements the conservation of score (COS) using the weighted rank approach algorithm (COS(WR)) developed recently by Akerman et al. (14). Specifically, our method searches for the existence of a defined consensus motif, ideally motifs that have been confirmed experimentally to bind known SFs. The list of motifs that were tested and verified is given in Table 1. The COS(WR) algorithm exploits two major attributes of functional binding sites on RNA: their preference to cluster in regulatory regions in order to enhance binding affinity and the tendency of the regulatory region to be conserved in evolution. COS(WR) scores are calculated in two steps: first, the weighted rank (WR) function is applied to calculate the clustering propensity of the motif; second, evolutionary conservation is estimated by further weighting the WR scores with the COS function. The COS function relaxes positional constraints imposed by sequence alignments, therefore it is ideal for estimating the evolutionary conservation of regulatory motifs. A detailed description of the algorithm is given in Akerman et al. (14).
Table 1.
List of SF-binding motifsa
| SF | Motifs |
|---|---|
| SF2ASF | crsmsgw, ugrwgvh |
| 9G8 | acgagagay, wggacra |
| SC35 | gryymcyr, ugcygyy |
| Tra2alpha | gaagaggaag |
| Tra2beta | gaagaa, ghvvganr, aaguguu |
| SRp20 | cuckucy, wcwwc |
| SRp40 | yywcwsg |
| SRp55 | yrcrkm |
| hnRNPA1 | uagaca, uagagu, uagggw |
| hnRNPAB | auagca |
| hnRNPH/F | ggcgg, gggug, uguggg, uugggu |
| MBNL | ygcuky |
| NOVA1 | ycay |
| PTB | cucucu, ucuu |
| CUG-BP | ugcug |
| YB1 | caaccacaa |
| FOX1/2 | ugcaug |
aThe SFs and their related motifs were extracted from the literature and are based on experimental data as detailed in Akerman et al. (14).
As described in Akerman et al. (14), the algorithm was tested on a set of experimentally validated sequences that bind specific factors and a control set of non-binding sites (56 and 502 sequences for positives and controls, respectively) and displayed a high 93% accuracy with a relatively low 1% false-positive rate. The results were obtained using a predefined set of parameters to calculate the WR function: the significant cut-off for the major hit, the suboptimal cut-off for the multiple hits and the window size. The COS(WR) algorithm performance was compared with the SF binding motif predictor ESEfinder (6,12), which is currently the only method available for mapping binding motifs of known SFs. As shown in Akerman et al. (14), the COS(WR) algorithm demonstrated significant higher specificity compared to ESEfinder motif predictions for the four SR proteins available in ESEfinder.
SFmap INPUT AND OUTPUT
The current SFmap version is designed for predicting and mapping SF binding sites in the HG. In addition, non-human RNA sequences are also accepted as input but will be processed without considering evolutionary conservation. As an input, SFmap requires a sequence or list of sequences in FASTA format that can be extracted from different assemblies of the HG. In addition, a user can choose to upload the sequence coordinates (chromosome: start-end:strand). Furthermore, the user is prompted to select the motifs of interest from a list of verified motifs (Table 1). As a default, all motifs stored in our database will be selected. Optionally, the user can add custom motifs, 4–10 nt-long, using IUPAC symbols. Custom motifs will be predicted by the same algorithm used to map other motifs in our database. To search for splicing motifs, the program runs the input sequence against the HG and retrieves the flanking sequences from the genome, as well as the human–mouse alignment of the region required for calculating the COS(WR) score. If no match is found, WR will be calculated for the central sequence excluding the terminal positions (i.e. window size/2 from each side). In cases where human–mouse alignments are not available, SFmap will employ the WR algorithm, which does not consider evolutionary conservation and thus does not require a sequence alignment. Notably, the user can deliberately choose to implement the WR algorithm that does not consider conservation information by manually selecting the WR box in the SFmap front page. This option will improve runtime, although at the expense of specificity. This option is obligatory when running non-human sequences. Since, for graphical display, SFmap uses the blat search algorithm (16) to determine input sequence coordinates in the HG, in cases where no match is found for the input sequence, results will be provided as a text file but will not be displayed visually.
SFmap outputs the predicted motifs and their calculated COS(WR) scores mapped to the input sequence. The results are provided both as a text file and in a visualized display of the motifs on the UCSC HG browser as a custom track. In order to define the significance of the score for each motif stored in our database, we calculated a threshold relative to a background model of random human sequences [for more details see (14)]. Furthermore, to accommodate motifs provided by the users, a theoretical threshold that best fits the experimental model was calculated. Finally, only motifs that were scored above the threshold are reported. For convenience, SFmap outputs the threshold calculated for each motif. The resulting text file includes a summary table of the motifs predicted by our algorithm and their location within the input sequence, including their genomic coordinates. A separate list including the position of the predicted motifs (hits) and the calculated score for each hit is given for each motif requested by the user. In cases where the input file includes multiple sequences, results are provided independently for each sequence (including a link to the text and the visualized display), as well as a summary file combining all motifs found in the input sequences. It is important to note that since the COS(WR) algorithm does not require 100% similarity of the binding site to the motif, different binding sites (related to the same SF or to different factors) can be predicted at the same genomic location.
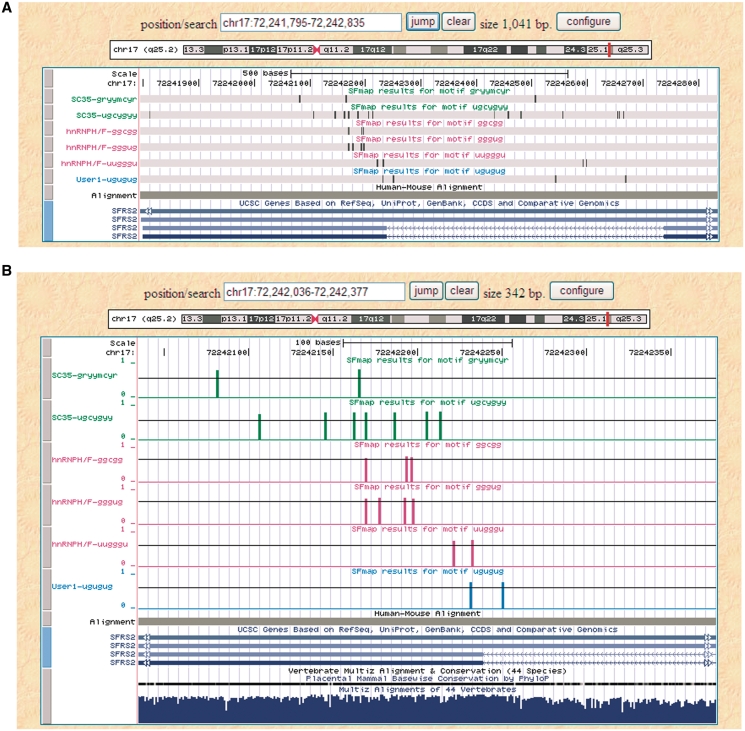
An example of a visualized display of SFmap in dense and full representation is shown in Figure 1A and 1B, respectively. In this example, the sequence of the 3′-untranslated region (UTR) of the SC35 gene was provided as the input to SFmap. SC35 is an SF from the SR protein family and was previously shown to be autoregulated by a cassette exon event at the 3′-UTR of its own gene (17). In a recent paper by Dreumont et al. (18), it was shown that two other SFs, hnRNP H and the TAR DNA-binding protein (TDP-43), antagonize the binding of SC35 to the junction between the retained intron and the terminal exon. In this example, we selected the two binding motifs of SC35 and the three hnRNP H/F motifs from the list of motifs provided by SFmap. In addition, we used the ‘new motif’ option to map the ‘UGUGUG motif’, which has been described in the literature as being the minimal binding motif of TDP-43 (19).
Figure 1.
SFmap output shown in a ‘dense’ (A) and a ‘full’ (B) format. In this example, SFmap was run on the 3′-UTR of the SC35 gene. The predicted sites for the motifs selected by the user are displayed on the UCSC HG Browser as a custom track. The top five motifs were selected from motifs that were stored in our database, and the last motif is a user-defined motif. As can be observed, most of the predicted binding sites of the three SFs are mapped to the intron/exon junction previously shown to be regulated by these factors (18).
As shown in Figure 1, the SF hits are displayed as a box (Figure 1A) or a thin bar (Figure 1B), designating the first position of the motif. In the ‘full’ display option (Figure 1B), bar height represents the score of the hit, which is detailed in the accompanying text output. Each row represents the results for a single motif specified on the left-hand side of the row. As indicated by arrows on the gene display (at the bottom of the figures), the SC35 gene is located on the antisense strand of the chromosome. Since the HG browser always displays the sense strand, the results in this example should be read from right to left. As illustrated in the figure, most of the binding sites of all three SFs were mapped to the junction region between the retained 3′-UTR intron and the 3′-UTR exon. Interestingly, when displaying the mammalian evolutionary conservation provided by the HG browser comparative genomic analysis track (bottom of Figure 1B), one can see that the entire 3′-UTR is highly conserved and by itself could not explain the specific localization of the binding sites of the three different SFs to the splicing junction. Overall, the results of SFmap are highly consistent with the experimental results presented in Dreumont et al. (18).
DISCUSSION
SFmap is designed to map SF binding sites in human genomic regions using the COS(WR) algorithm (14). The COS(WR) algorithm takes two important features of SF binding sites into account: their propensity to cluster and enhance binding affinity and specificity; and the tendency of the genomic environment of the binding site to be conserved. Taking these features into consideration, the method was shown to perform very accurately with a relatively low false-positive rate (14). The method was tested on a fixed set of experimentally defined binding motifs, however, it can be applied to any other motif of interest. Thus, though SFmap was specifically designed to map SF binding sites, it could potentially be applied to map binding sites of other RNA binding protein motifs on the RNA that are assumed to bind in a similar fashion, such as the Pumilio RNA-binding protein, which was recently shown to bind the 3′-UTR in a cooperative manner (20). Furthermore, the main advantage of the algorithm is that it derives standardized scores for any given motif so that predictions of different binding sites are comparable. Nevertheless, it is very important to note that the COS(WR) score does not reflect the binding affinity or functionality of the site. Moreover, since the COS(WR) algorithm has been designed to identify degenerative binding motifs relying on compensating information from the sequence environment, it is likely that in some cases, SFmap will map different binding motifs to the same genomic position. Even though different motifs will most likely yield a different score (both above the motif threshold), one should be very careful when selecting the best scored motif. Notably, the existence of two SF hits in the same position could indicate competition between two factors, such as that reported in the splicing regulation of the 3′-UTR of the SC35 gene (18) shown in the example in Figure 1. Moreover, we would like to emphasize that COS(WR) may not be suitable for uniquely indentifying binding sites of proteins that have very stringent binding preferences. In the latter cases, we expect that SFmap will be sensitive but not highly selective, and thus may obtain a higher number of false-positive results with relatively high scores. It is important to emphasize that though the COS(WR) algorithm was tested on a set of 30 different experimentally verified motifs, we cannot fully anticipate prediction accuracy for motifs that have not been tested. Thus, though SFmap can direct researchers to regions in the sequences that are likely to bind SFs or other RNA-binding proteins, direct experimental evidence will still be required to confirm the predictions.
CONCLUSIONS
In the post-genomic era, tools for mapping SF binding sites are in great need, both for basic research and for the detection of disease-associated mutations in RNA regulatory sequences. The SFmap server, which implements the COS(WR) algorithm (14), has been designed for mapping binding sites of known proteins given their consensus motifs. Since the method is very general, it can be applied for predicting the motifs of other RNA-binding proteins that are expected to have similar binding preferences. Once more detailed information on binding motifs is revealed from the experimental data, SFmap could be implemented to enhance high accurate genome-wide predictions of binding sites of SFs in particular and RNA-binding proteins in general.
FUNDING
Elyahu Pen Research Fund; the Israeli Science Foundation (grant number 1297/09 to Y.M.G.). Funding for open access charge: The Israeli Science Foundation (grant number 1297/09 to Y.M.G.).
Conflict of interest statement. None declared.
ACKNOWLEDGEMENT
We would like to thank the many users for their useful comments and suggestions for improving the web site.
REFERENCES
- 1.Chen M, Manley JL. Mechanisms of alternative splicing regulation: insights from molecular and genomics approaches. Nat. Rev. Mol. Cell. Biol. 2009;10:741–754. doi: 10.1038/nrm2777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cooper TA, Wan L, Dreyfuss G. RNA and disease. Cell. 2009;136:777–793. doi: 10.1016/j.cell.2009.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schwartz S, Hall E, Ast G. SROOGLE: webserver for integrative, user-friendly visualization of splicing signals. Nucleic Acids Res. 2009;37:W189–W192. doi: 10.1093/nar/gkp320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang Z, Burge CB. Splicing regulation: from a parts list of regulatory elements to an integrated splicing code. RNA. 2008;14:802–813. doi: 10.1261/rna.876308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ladd AN, Cooper TA. Finding signals that regulate alternative splicing in the post-genomic era. Genome Biol. 2002;3 doi: 10.1186/gb-2002-3-11-reviews0008. reviews0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cartegni L, Wang J, Zhu Z, Zhang MQ, Krainer AR. ESEfinder: a web resource to identify exonic splicing enhancers. Nucleic Acids Res. 2003;31:3568–3571. doi: 10.1093/nar/gkg616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fairbrother WG, Yeo GW, Yeh R, Goldstein P, Mawson M, Sharp PA, Burge CB. RESCUE-ESE identifies candidate exonic splicing enhancers in vertebrate exons. Nucleic Acids Res. 2004;32:W187–W190. doi: 10.1093/nar/gkh393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang XH, Kangsamaksin T, Chao MS, Banerjee JK, Chasin LA. Exon inclusion is dependent on predictable exonic splicing enhancers. Mol. Cell Biol. 2005;25:7323–7332. doi: 10.1128/MCB.25.16.7323-7332.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang Z, Rolish ME, Yeo G, Tung V, Mawson M, Burge CB. Systematic identification and analysis of exonic splicing silencers. Cell. 2004;119:831–845. doi: 10.1016/j.cell.2004.11.010. [DOI] [PubMed] [Google Scholar]
- 10.Goren A, Ram O, Amit M, Keren H, Lev-Maor G, Vig I, Pupko T, Ast G. Comparative analysis identifies exonic splicing regulatory sequences–the complex definition of enhancers and silencers. Mol. Cell. 2006;22:769–781. doi: 10.1016/j.molcel.2006.05.008. [DOI] [PubMed] [Google Scholar]
- 11.Chasin LA. Searching for splicing motifs. Adv. Exp. Med. Biol. 2007;623:85–106. doi: 10.1007/978-0-387-77374-2_6. [DOI] [PubMed] [Google Scholar]
- 12.Smith PJ, Zhang C, Wang J, Chew SL, Zhang MQ, Krainer AR. An increased specificity score matrix for the prediction of SF2/ASF-specific exonic splicing enhancers. Hum. Mol. Genet. 2006;15:2490–2508. doi: 10.1093/hmg/ddl171. [DOI] [PubMed] [Google Scholar]
- 13.Huang HY, Chien CH, Jen KH, Huang HD. RegRNA: an integrated web server for identifying regulatory RNA motifs and elements. Nucleic Acids Res. 2006;34:W429–W434. doi: 10.1093/nar/gkl333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Akerman M, David-Eden H, Pinter RY, Mandel-Gutfreund Y. A computational approach for genome-wide mapping of splicing factor binding sites. Genome Biol. 2009;10:R30. doi: 10.1186/gb-2009-10-3-r30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kuhn RM, Karolchik D, Zweig AS, Wang T, Smith KE, Rosenbloom KR, Rhead B, Raney BJ, Pohl A, Pheasant M, et al. The UCSC Genome Browser Database: update 2009. Nucleic Acids Res. 2009;37:D755–D761. doi: 10.1093/nar/gkn875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kent WJ. BLAT–the BLAST-like alignment tool. Genome Res. 2002;12:656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sureau A, Gattoni R, Dooghe Y, Stevenin J, Soret J. SC35 autoregulates its expression by promoting splicing events that destabilize its mRNAs. EMBO J. 2001;20:1785–1796. doi: 10.1093/emboj/20.7.1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dreumont N, Hardy S, Behm-Ansmant I, Kister L, Branlant C, Stevenin J, Bourgeois CF. Antagonistic factors control the unproductive splicing of SC35 terminal intron. Nucleic Acids Res. 2009;38:1353–1366. doi: 10.1093/nar/gkp1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Buratti E, Baralle FE. Multiple roles of TDP-43 in gene expression, splicing regulation, and human disease. Front. Biosci. 2008;13:867–878. doi: 10.2741/2727. [DOI] [PubMed] [Google Scholar]
- 20.Gupta YK, Lee TH, Edwards TA, Escalante CR, Kadyrova LY, Wharton RP, Aggarwal AK. Co-occupancy of two Pumilio molecules on a single hunchback NRE. RNA. 2009;15:1029–1035. doi: 10.1261/rna.1327609. [DOI] [PMC free article] [PubMed] [Google Scholar]