Abstract
The GlycoViewer (http://www.systemsbiology.org.au/glycoviewer) is a web-based tool that can visualize, summarize and compare sets of glycan structures. Its input is a group of glycan structures; these can be entered as a list in IUPAC format or via a sugar structure builder. Its output is a detailed graphic, which summarizes all salient features of the glycans according to the shapes of the core structures, the nature and length of any chains, and the types of terminal epitopes. The tool can summarize up to hundreds of structures in a single figure. This allows unique, high-level views to be generated of glycans from one protein, from a cell, a tissue or a whole organism. Use of the tool is illustrated in the analysis of normal and disease-associated glycans from the human glycoproteome.
INTRODUCTION
Glycomics is the study of all glycans from a cell, tissue or system. Large numbers of glycan structures are frequently reported in research manuscripts, predominantly determined by mass spectrometry. Increasingly, these are captured in structure repositories and high-quality curated databases, such as KEGG glycan, GlycomeDB and GlycoSuiteDB (1–3). Whilst there are thousands of structures in these databases, it has been challenging to combine and visualize these data in a simple fashion. This has prevented the generation of holistic views of the glycome, at the level of a cell, tissue or whole organism. Current depictions of glycans, including the Oxford and Consortium for Functional Glycomics (CFG) schema, are tailored to represent single 2D structures and are not designed to describe families of structures. KEGG composite structure maps (1) summarize structure and pathway data, but are challenging to interpret. Furthermore, they are not designed to compare two or more sets of glycan structures or those from different levels of the glycome. Here we describe the GlycoViewer tool, a means by which the glycome is given a single representation. This provides a unique, high-order view, permitting global analysis and comparison of glycomic data.
MATERIALS AND METHODS
To build a single representation of the glycome, the features contributing most to the variation between structures had to be defined. An evaluation of all human glycan structures from GlycomeDB (2), in the context of known glycosylation pathways, revealed that N- and O-linked structures could be categorized using just three criteria: (i) the type and shape of the core structure, (ii) the nature and length of any chain, and (iii) the nature of any terminal epitopes (e.g. sialylation, A or B antigen). The relationships between these criteria were also captured.
To summarize a set of glycan structures, these criteria are applied systematically. Each input structure is traversed from the reducing terminus to non-reducing termini and each of the criteria, above, are evaluated against each of the residues. A decision is made to display, annotate or ‘compress’ each of the residues. Statistics describing the number of structures that have particular features (e.g. chain types or terminal epitopes) are calculated. Structures from any set that appear incomplete or erroneous, which are inconsistent with the criteria, are removed. The final high confidence set of structures is used to build a composite structure, from the union of all supplied structures. Separate composite structures are built for N- and O-linked sugars. To visualize these composite structures, a modified CFG schema is used to show the criteria of shape, nature and length, and terminal epitopes. Annotations to represent the statistics are also built into the graph. Histograms to quantify branching are shown alongside, together with names of any branch types.
The summarizing process has been built into the GlycoViewer tool (http://www.systemsbiology.org.au/glycoviewer). Lists containing up to hundreds of structures can be submitted, for example from databases such as GlycomeDB or GlycoSuiteDB (2,3). Structures must adhere to IUPAC nomenclature. Alternatively, a structure builder is supplied so lists can be constructed as required and then analysed. The tool is freely available and has no login requirement. Detailed instructions on the interpretation of the tool’s output are given on the web site, on the page titled ‘Interpreting the Output’ and are given here as Supplementary Data.
USAGE
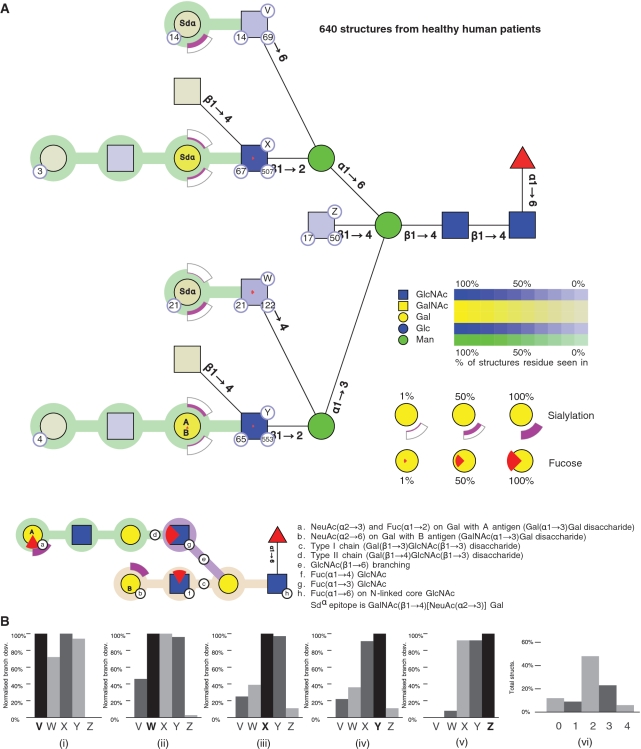
We give two examples to show how the GlycoViewer can summarize, analyse and compare glycomic data. As a first example, the tool was used to analyse all known human N-linked structures from the glycome of healthy patients, as documented in the GlycoSuite database (3). These structures were obtained by retrieving all 3183 structures for Homo sapiens and filtering to remove structures associated with disease, a recombinant system or cell line and those which were O-linked. Figure 1 summarizes 640 structures to show all monosaccharides present, branching properties of the N-linked structures, lengths of each chain and the degree of sialylation and fucosylation. Qualitative features of the glycome are given and semi-quantitative measurements show the frequency of particular residues in structures. The representation highlights specific features of large sets of structures. For the 640 structures, it is easily seen that all chain extensions are Type II, with the majority of structures being bi-antennary (labelled X and Y in Figure 1). The bisecting GlcNAc (labelled Z) is mutually exclusive to a β1–6 GlcNAc (labelled V) linked to the Man α1–6 branch. The latter feature has recently been reported elsewhere (4) and has been attributed to β1–4 N-acetylglucosaminyltransferase III suppressing β1–4 N-acetylglucosaminyltransferase V. The absence of β1-4 linked GlcNAc on the Man α1–6 branch of the N-linked core, also clear in the figure, has also been previously noted (5). The GlycoViewer enabled these features to be discerned in a single graphic; a one-by-one analysis of the entire structure set would have been laborious, and may not have provided the same high-level insights.
Figure 1.
The composite structure for 640 N-glycans, derived from human tissue. (A) Residues in all structures have CFG icons and colours with saturation of colour representing the abundance of a residue in the set of structures. Residues may be annotated to show the number of structures in the database containing a residue (number in bottom right circle), number of publications reporting the residue in the structure (number in bottom left circle) and a label (V, W, X, Y, Z) that identifies a branch (capital letter in top right circle). Linkages between core residues are labelled with anomer and ring substitution position whereas linkages in chains have coloured linking connections (explained in the key to the figure). Terminal epitopes upon chains are represented using arcs (NeuAc) and wedges (Fucose). Arcs show the degree of sialylation of a residue in a set of structures; the degree of fill in the arc correlates with the degree of sialylation; the two possible positions of sialylation (α2–3 and α2–6) are positioned below and above the residue, respectively. Wedges show the degree of fucosylation, with full fucosylation represented by a full wedge. A downward-facing wedge is a fucose in an α1–2 linkage, a left-facing wedge represents α1–3 linkage and an upward facing wedge represents an α1–4 linkage. Core fucosylation is shown by a standard symbol. The annotation inside the residues of a chain (e.g. Sdα, A or B) indicate terminal epitopes comprised of Gal and GalNAc residues. (B) Histograms measuring the overall branching characteristics of the set of glycan structures. One histogram is shown per labelled branch (i–v), with each histogram comparing the number of times other branches occur with respect to that branch. Thus the histogram for branch V (i) shows that structures containing branch V can contain branches W–Y but do not contain branch Z. The overall degree of branching in the set of glycans is shown in a second type of histogram (vi), where the proportion of structures in the set containing 0, 1…, n branches is graphed.
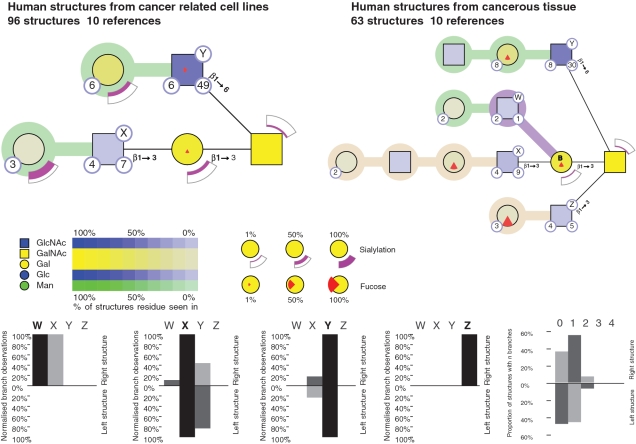
As a second example, sets of O-linked glycans from two biological states are compared: those from cancerous human tissue (98 structures) and human cancer cell lines (63 structures). The tool facilitates a side by side analysis, with two summary representations (Figure 2). Most striking is the presence of Type I chains on tissue-derived glycoproteins and their complete absence in cell line-derived glycoproteins. Tissue-derived proteins also show increased fucosylation and concomitant decrease in sialylation as compared to cell line-derived glycans. Our tool also revealed other features of the tissue-derived glycans. The presence of the core β1–3 GlcNAc (labelled Z in Figure 2) is mutually exclusive to the presence of core β1–6 GlcNAc (labeled Y), precluding the existence of core IV structures. Core β1–3 GlcNAc (Z) is also mutually exclusive to the presence of the core β1–3 Gal (W,X); this is to be expected as they share the same linkage with the core GalNAc. It is interesting to note that this approach, which facilitated the comparison of two biological states, could also be used to compare the glycans from two or more proteins or even the glycome from two or more species.
Figure 2.
Comparison of O-linked glycan structures from human cancer cell lines (98 structures) and cancerous human tissues (63 structures). This comparison was done by entering the two sets of structures, as list 1 and list 2, and using the ‘compare list’ function of GlycoViewer. Note that in this figure, the histograms compare the overall branching in the two sets of glycans. Bars going up represent branching in the human cancer tissues; bars going down represent branching in cancer-related cell lines.
DISCUSSION AND CONCLUSION
The GlycoViewer tool, described here, helps reveal the complexities of the glycome. It allows small or large sets of glycan structures to be analysed and compared. Results are reported in a single, comprehensive figure. Qualitative and semi-quantitative aspects are highlighted in the context of structures and graphed in a series of analytical histograms. We believe the tool can form a logical endpoint in an analysis pipeline for glycomic data and has potential to provide holistic insights that are otherwise difficult to discern. It provides a single, common interface to the glycome at the level of the cell, the tissue or entire organism. One major consideration in the use of our technology, and similar approaches that examine large sets of structures, is the quality of input data. Whilst there are numerous databases of glycan structures in use, the quality of structures in each database and the biological descriptors are variable. In part, this reflects the challenge of curating structural information, the difficulties in unambiguous determination of structures even by mass spectrometry and the need for an international co-ordinated effort in this regard. In the analyses presented here, we used structures from the GlycoSuiteDB database (3), recently released into the public domain (http://glycosuitedb.expasy.org). This is one of the few databases which represent structures in IUPAC format. Whilst this database does not contain a complete glycome for any species, it is sufficiently diverse and curated to allow complex queries and analyses to be made and high-level features of glycome to be discerned. The efficacy and utility of the GlycoViewer tool will improve as glycan structure databases become increasingly complete.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
NSW Science State Leveraging Fund, the University of New South Wales and the EU (6th Research Framework Program, RIDS contract number 011952). Funding for open access charge: New South Wales State Government Science Leveraging Funds and the University of New South Wales.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Simone Li for programming, Adrian Plummer for IT support and for hosting of this tool, and Tyrian Diagnostics for the release of GlycoSuiteDB to the public domain.
REFERENCES
- 1.Hashimoto K, Kawano S, Goto S, Aoki-Kinoshita KF, Kawashima M, Kanehisa M. A global representation of the carbohydrate structures: a tool for the analysis of glycan. Genome Inform. 2005;16:214–222. [PubMed] [Google Scholar]
- 2.Ranzinger R, Herget S, Wetter T, von der Lieth CW. GlycomeDB – integration of open-access carbohydrate structure databases. BMC Bioinformatics. 2008;9:384. doi: 10.1186/1471-2105-9-384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cooper CA, Joshi HJ, Harrison MJ, Wilkins MR, Packer NH. GlycosuiteDB : a curated relational database of glycoprotein glycan structures and their biological sources. Nucleic Acids Res. 2003;31:511–513. doi: 10.1093/nar/gkg099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gu J, Sato Y, Kariya Y, Isaji T, Taniguchi N, Fukuda T. A mutual regulation between cell-cell adhesion and N-glycosylation: implication of the bisecting GlcNAc for biological functions. J. Proteome Res. 2009;8:431–435. doi: 10.1021/pr800674g. [DOI] [PubMed] [Google Scholar]
- 5.Brockhausen I, Hull E, Hindsgaul O, Schachter H, Shah RN, Michnick SW, Carver JP. Control of glycoprotein synthesis: detection and characterization of a novel branching enzyme from hen oviduct, UDP-N-acetylglucosamine:GlcNAc beta 1-6 (GlcNAc beta 1-2)Man alpha-R (GlcNAc to Man) beta-4-N-acetylglucosaminyltransferase VI. J. Biol. Chem. 1989;264:11211–11221. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.