Abstract
HexServer (http://hexserver.loria.fr/) is the first Fourier transform (FFT)-based protein docking server to be powered by graphics processors. Using two graphics processors simultaneously, a typical 6D docking run takes ∼15 s, which is up to two orders of magnitude faster than conventional FFT-based docking approaches using comparable resolution and scoring functions. The server requires two protein structures in PDB format to be uploaded, and it produces a ranked list of up to 1000 docking predictions. Knowledge of one or both protein binding sites may be used to focus and shorten the calculation when such information is available. The first 20 predictions may be accessed individually, and a single file of all predicted orientations may be downloaded as a compressed multi-model PDB file. The server is publicly available and does not require any registration or identification by the user.
INTRODUCTION
Protein docking is the task of calculating the 3D structure of a protein complex from its unbound or model-built subunits. Although proteins are intrinsically flexible, many protein docking algorithms begin by assuming that the proteins are rigid and they use geometric hashing (1) or fast Fourier transform (FFT) correlation techniques (2) to find a relatively small number of putative docking orientations which may be refined and re-scored using more sophisticated techniques.
In recent years, several protein docking programs have been made available as web servers. These range from the rapid PatchDock server (3), which is based on a rigid-body geometric hashing algorithm (4), to much more computationally intensive approaches incorporating models of flexibility such as RosettaDock (5) and Haddock (6). Several FFT-based docking programs have also been made available as web servers e.g. ClusPro (7), GRAMM-X (8) and ZDOCK (9). Like the geometric hashing approach, the FFT-based approaches assume that the proteins to be docked are rigid, but they sample densely all possible rigid-body orientations in the 6D search space. However, because most FFT-based approaches use 3D Cartesian grid representations of the proteins, they can only compute translational correlations, and these must be repeated over multiple rotational samples in order to cover the 6D search space. Thus, despite the rigid-body assumption, Cartesian grid-based FFT docking algorithms are inherently computationally expensive.
In order to address the main limitations of the Cartesian FFT approaches, we developed the ‘Hex’ spherical polar Fourier (SPF) approach which uses rotational correlations (10), and which reduces execution times to a matter of minutes (11). Nonetheless, we recently adapted the Hex algorithm to obtain a further significant speed-up by exploiting the enormous computational power of modern graphics processor units (GPUs; in preparation) using the CUDA (Common Unified Device Architecture) development tools (http://www.nvidia.com/object/cuda_home.htm). For typical Hex docking calculations, a single high-performance GPU can evaluate ∼170 million trial orientations/second. This corresponds to a speed-up of at least a factor of 45 compared to a contemporary central processor unit (CPU), and which is up to two orders of magnitude faster than conventional Cartesian grid-based FFT docking approaches. However, because high performance GPUs are relatively expensive, we have developed HexServer, a web interface for Hex, in order to make our GPU-accelerated docking approach widely and freely available.
The Hex SPF algorithm has been validated in the CAPRI (Critical Assessment of PRedicted Interactions) blind docking experiment (12), and an acceptable rigid-body Hex prediction has often been found within the top 100 orientations in recent CAPRI scoring sections. Thus, HexServer provides a very fast and convenient way to generate high quality docking predictions for subsequent refinement.
MATERIALS AND METHODS
System architecture
HexServer is implemented using a small number of web pages and shell scripts which communicate via a MySQL (http://www.mysql.com/) database. The computational part of the server consists of a 32-node cluster running the CentOS 5.2 operating system and using the OAR batch scheduling system (http://oar.imag.fr/). Each node consists of two quad-core Intel Xeon 2.5 GHz CPUs, and eight of the nodes are equipped with two Nvidia Tesla C1060 GPUs. Hence, a total of 256 CPU cores and 16 GPUs are currently available on our server.
The web interface is implemented using the PHP scripting language, through which the user's PDB files are uploaded and stored in a MySQL database along with the other parameters of the docking job. Each job is identified by a unique 13 digit job number. The web interface performs some basic sanity checks on the input data in order to highlight errors quickly and to avoid wasting processor time on the server. A Linux shell script running on the compute server periodically polls the databases for new jobs. When a new job is found, another shell script is generated and is submitted to the OAR batch queuing system. This script executes the Hex program and copies the results files back to the MySQL database when the job has finished. The submitting web page periodically polls the database for an indication that the job has completed, and the results may be accessed from a job-specific web link generated automatically using the job number. If the user has provided an e-mail address, he will be sent a mail message containing a link to the job's results page.
HexServer does not require any kind of user registration or identification, and all results are accessible using only the 13 digit job number. All PDB coordinate files of each job are deleted from the system after 24 h, although other details of the job are stored for statistical purposes.
Inputs
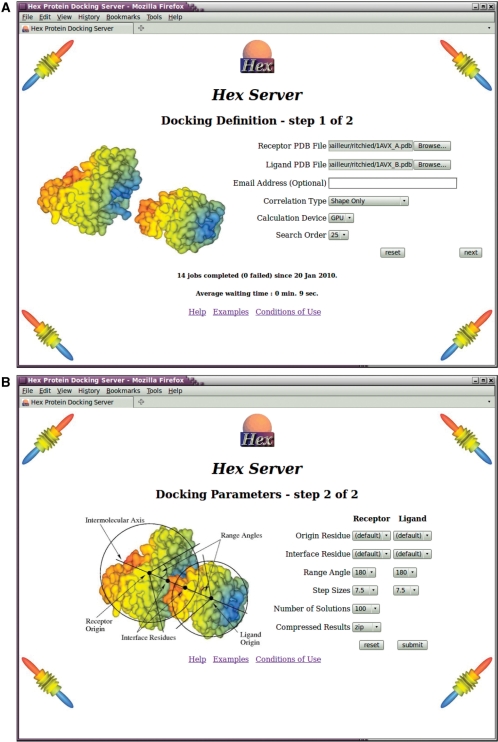
HexServer has an easy-to-use form-based interface, through which users may upload a pair of protein structures in PDB format. Users may optionally provide an e-mail address for notification of the status of their jobs. Figure 1 shows the web interface for defining the parameters of a docking job. For a blind unconstrained 6D docking run, it is normally sufficient to use the default values for all parameters. If the proteins to be docked have large and opposite formal charges, or if electrostatic interactions are known to be important, it is often beneficial to request a shape plus electrostatic calculation. Otherwise, a shape-only correlation is recommended.
Figure 1.
Screenshots of the two dataentry web pages of the HexServer interface. (A) Top: the first web page is used to specify the PDB files to be uploaded, and the type of docking calculation to be performed. (B) Bottom: the second web page may be used to define optional interface residues and angular search ranges to focus the search around a known or hypothesized interface. By convention, the larger of the two proteins is called the ‘receptor’ and the smaller is called the ‘ligand,’ although Hex treats the two proteins equally. All input parameters are explained in further detail in the online Help page, and some typical protein domains are available from the ‘Examples’ page.
As described previously (11), all Hex docking correlations use SPF shape–density representations to polynomial order 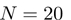 in order to generate very rapidly a list of up to 25 000 candidate solutions. We find that the top 3000 orientations nearly always include some near-native orientations but a larger list is used to avoid pruning good candidates in exceptional cases. These candidate solutions are then re-scored using higher order shape-only or shape plus electro-static correlations (using e.g. polynomials to order
in order to generate very rapidly a list of up to 25 000 candidate solutions. We find that the top 3000 orientations nearly always include some near-native orientations but a larger list is used to avoid pruning good candidates in exceptional cases. These candidate solutions are then re-scored using higher order shape-only or shape plus electro-static correlations (using e.g. polynomials to order 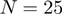 or
or 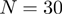 ), as selected by the user. Requesting polynomial order
), as selected by the user. Requesting polynomial order 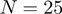 (the default) gives relatively soft representations of each protein whereas order
(the default) gives relatively soft representations of each protein whereas order 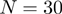 polynomials give somewhat sharper representations.
polynomials give somewhat sharper representations.
If prior information is available about one or both binding sites, the user can request that the docking search will be focused around a selected interface residue on one or both docking partners. As illustrated in Figure 1B, this is achieved by specifying one central residue from each protein to define an intermolecular axis, and by specifying two further residues to be placed on the intermolecular axis near the protein–protein interface. The user may then specify an angular search range (e.g. of 45°) for each protein with respect to the intermolecular axis in order to constrain the rotational search around the putative interface.
Outputs
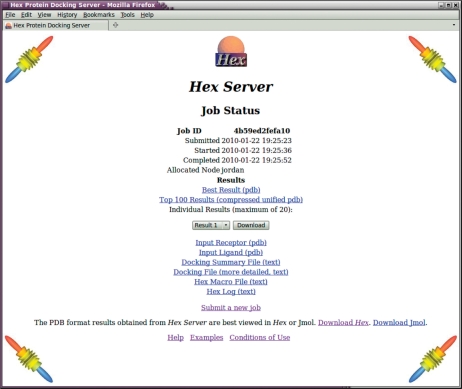
Once a job is complete, the user is directed to a simple results page (Figure 2) where he may download a ranked list of predicted complexes. Because HexServer aims to provide a relatively large number of putative complexes for re-scoring, the requested number of predictions is presented as a single compressed multi-structure PDB file in which each structure is identified using the standard PDB ‘MODEL’ and ‘ENDMDL’ keywords. This file may be requested in any of the ‘zip’, ‘gzip’ or ‘bzip2’ compression formats. The first 20 structures are also made available individually in uncompressed PDB format. Thus the user may quickly preview the predictions before downloading the large multi-structure results file.
Figure 2.
A screenshot of a results page generated by HexServer.
RESULTS AND DISCUSSION
Overall performance
The HexServer web interface provides a simple and easy way to prepare protein–protein docking calculations using Hex. The computational backend provides public access to a powerful GPU-based cluster. The OAR batch queuing system ensures that only one docking job can execute at a time on each pair of GPUs, which maximizes job throughput and avoids contention for resources. On our system, a typical exhaustive 6D rigid-body docking search takes around 15 s when using two C1060 GPUs simultaneously. If knowledge of even just one interface residue from one or both proteins is available, it can be used very effectively to constrain the docking search around the known or supposed interface. This further reduces the overall docking time, and significantly improves the quality of the predicted complexes (11).
Comparison with ZDOCK
In order to illustrate the speed-up given by our GPU-accelerated approach over a conventional Cartesian grid-based FFT docking calculation, we docked the PDB structures given on the HexServer ‘Examples’ page (porcine trypsin and soybean trypsin inhibitor, PDB code 1AVX) using Hex 6.0, HexServer (which invokes Hex 6.0), ZDOCK 3.0.1 and the corresponding ZDOCK server (http://zdock.bu.edu/).
It should be noted, however, that it is difficult to make an exact comparison due to the fundamental difference in how the search space is partitioned in the SPF (five rotations and one translation) and Cartesian (three rotations and three translations) coordinate systems, and because of other differences in the orientational sampling techniques used. It is also worth emphasizing that the speed of any FFT-based approach depends critically on the sampling resolution used: doubling the step size in each dimension will give a speed-up of 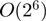 , but using large step sizes entails a risk that good solutions will be missed. Hence, both Hex and ZDOCK employ a strategy of densely sampling the search space and then clustering solutions with similar orientations. For example, by default, both Hex and HexServer use 64 steps of 5.625° for rotational increments about the intermolecular axis, and they use icosahedral tessellations of the sphere of 812 vertices to give angular rotation samples of about 7.5 for the remaining angular degrees of freedom. Hence, the default sampling density in Hex (812×64=51 968 ligand rotations) corresponds quite closely to ZDOCK's ‘dense’ sampling mode (54 000 ligand rotational steps of about 6°). On the other hand, ZDOCK server does not offer dense sampling due to its high computational cost, and instead uses the default ZDOCK coarse sampling level of 15° (3600 ligand rotations). Furthermore, Hex and HexServer use translational steps of 0.8Å, which is somewhat finer than the 1.2Å grid spacing used in ZDOCK and ZDOCK server (9).
, but using large step sizes entails a risk that good solutions will be missed. Hence, both Hex and ZDOCK employ a strategy of densely sampling the search space and then clustering solutions with similar orientations. For example, by default, both Hex and HexServer use 64 steps of 5.625° for rotational increments about the intermolecular axis, and they use icosahedral tessellations of the sphere of 812 vertices to give angular rotation samples of about 7.5 for the remaining angular degrees of freedom. Hence, the default sampling density in Hex (812×64=51 968 ligand rotations) corresponds quite closely to ZDOCK's ‘dense’ sampling mode (54 000 ligand rotational steps of about 6°). On the other hand, ZDOCK server does not offer dense sampling due to its high computational cost, and instead uses the default ZDOCK coarse sampling level of 15° (3600 ligand rotations). Furthermore, Hex and HexServer use translational steps of 0.8Å, which is somewhat finer than the 1.2Å grid spacing used in ZDOCK and ZDOCK server (9).
It should also be noted that Hex and ZDOCK employ different scoring functions. For example, Hex calculates an excluded volume model of shape complementarity with an optional in vacuo electrostatic contribution (10), whereas ZDOCK uses a scoring function composed from shape, electrostatics and an atomic contact model of desolvation (9). Hence, the two programs will inevitably produce different lists of predictions, although the overall computational complexity of their scoring functions is broadly similar.
Bearing the above observations in mind, Table 1 shows that for dense sampling, using Hex with one high performance GPU is about 330 times faster than using ZDOCK on a single 2.5 GHz CPU core. Although 15° sampling is not publicly available in HexServer due to the risk of missing good solutions, performing such coarse sampling using two GPUs takes only around 3s, which is around 160 times faster than the corresponding calculation on the ZDOCK server. These figures justify our claim that using GPUs to accelerate Hex docking calculations can be up to two orders of magnitude faster than conventional FFT-based docking approaches when using comparable search resolutions and scoring functions.
Table 1.
Timing comparisons of Hex and ZDOCKa
| Modeb | Hex | Hex | ZDOCK | HexServer | ZDOCK server |
|---|---|---|---|---|---|
| (CPUc) | (GPUd) | (CPUc) | (GPUe) | (CPUf) | |
| dense | 240 | 22 | 7255 | 15 | – |
| coarse | 52 | 5 | 500 | 3 | 900 |
aAll times are given in seconds: the server timings exclude any networking delays or time spent waiting in a queue.
bHere, a ‘coarse’ sampling mode corresponds to 15° angular steps (about 3600 ligand rotations), whereas ‘dense’ sampling corresponds to 6° angular steps (about 54 000 ligand rotations). Dense sampling is not available in ZDOCK server.
cUsing 3D FFTs on one 2.5 GHz Intel Xeon processor.
dUsing 1D Hex FFTs on one Nvidia C1060 processor.
eUsing 1D Hex FFTs on two Nvidia C1060 processors.
fUsing 3D FFTs on eight IBM 1.1 GHz p655 processors.
Recommendations for use
Although docking programs such as Hex and ZDOCK can often produce near-native orientations within the first few hundred predictions, it remains a significant challenge to identify which orientations are in fact the near-native ones. We therefore recommend that users should visualize docking predictions from HexServer using an interactive graphics tool such as Jmol (http://jmol.sourceforge.net/download/), VMD (http://www.ks.uiuc.edu/Research/vmd/) or indeed the stand-alone version of Hex itself (http://hex.loria.fr/). If biological knowledge about the interaction is available, this should be used to colour-code known interaction residues to help assess each orientation. It is also recommended to consider refining selected orientations using short molecular dynamics runs or by submitting them to a flexible docking server such as RosettaDock (5) or Haddock (6).
In principle, PDB files could be loaded into HexServer directly from the PDB repository (http://www.rcsb.org). However, we recommend that the user first download and examine the protein structures to be docked, because it is often necessary to delete unwanted domains and hetero groups before performing a docking calculation.
It should be noted that Hex is designed for docking typical protein domains of up to around 150 amino acid residues. To dock proteins which are larger than this, it is recommended to perform a constrained angular search with respect to an explicitly specified initial orientation, as described above.
In order to keep the web interface simple, many of the more advanced or specialized features in Hex are not available in HexServer. Hence, we encourage users both to experiment with HexServer and to download the Hex program. Binary executables are available for several versions of popular operating systems, of which an increasing number support CUDA-based GPUs.
CONCLUSION
HexServer provides a convenient way to perform exhaustive GPU-accelerated FFT-based rigid-body docking predictions without requiring the user to invest in special hardware. Each blind 6D docking calculation takes only ∼15s on our server, and the results for each docking run are accessible for up to 24 h from a unique web page. Thus, users may quickly and easily obtain a list of high quality docking predictions for subsequent refinement and analysis.
FUNDING
Part of this work was funded by Agence Nationale de la Recherche (grant reference ANR-08-CEXC-017-01). The compute cluster is co-funded by INRIA and Region Lorraine. Funding for open acess charge: Agence Nationale de la Recherche.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We thank Birama Ndiaye and Olivier Demengeon for assistance with configuring OAR and the compute cluster.
REFERENCES
- 1.Bachar O, Fischer D, Nussinov R, Wolfson HJ. A computer vision based technique for 3D sequence-independent structural comparison of proteins. Protein Eng. 1993;6:279–288. doi: 10.1093/protein/6.3.279. [DOI] [PubMed] [Google Scholar]
- 2.Katchalski-Katzir E, Shariv I, Eisenstein M, Friesem AA, Aflalo C, Vakser IA. Molecular surface recognition: determination of geometric fit between proteins and their ligands by correlation techniques. Proc. Natl Acad. Sci. USA. 1992;89:2195–2199. doi: 10.1073/pnas.89.6.2195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schneidman-Duhovny D, Inbar Y, Nussinov R, Wolfson HJ. PatchDock and SymmDock: servers for rigid and symmetric docking. Nucleic Acids Res. 2005;33:W363–W367. doi: 10.1093/nar/gki481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Duhovny D, Nussinov R, Wolfson HJ. Proceedings of the 2nd Workshop on Algorithms in Bioinformatics (WABI), Lecture Notes in Computer Science 2452. Berlin: Springer; 2002. Efficient unbound docking of rigid molecules; pp. 185–200. [Google Scholar]
- 5.Lyskov S, Gray JJ. The RosettaDock server for local protein-protein docking. Nucleic Acids Res. 2008;36:W233–W238. doi: 10.1093/nar/gkn216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dominguez C, Boelens R, Bonvin A.M.JJ. HADDOCK: a protein-protein docking approach based on biochemical or biophysical information. J. Am. Chem. Soc. 2003;125:1731–1737. doi: 10.1021/ja026939x. [DOI] [PubMed] [Google Scholar]
- 7.Comeau SR, Gatchell DW, Vajda S, Camacho CJ. ClusPro: a fully automated algorithm for protein-protein docking. Nucleic Acids Res. 2004;32:W96–W99. doi: 10.1093/nar/gkh354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tovchigrechko A, Vakser IA. GRAMM-X public web server for protein-protein docking. Nucleic Acids Res. 2006;34:W310–W314. doi: 10.1093/nar/gkl206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen R, Li L, Weng Z. ZDOCK: an initial-stage protein-docking algorithm. Proteins: Struct. Func. Bioinf. 2003;52:80–87. doi: 10.1002/prot.10389. [DOI] [PubMed] [Google Scholar]
- 10.Ritchie DW, Kemp G.JL. Protein docking using spherical polar Fourier correlations. Proteins: Struct. Func. Genet. 2000;39:178–194. [PubMed] [Google Scholar]
- 11.Ritchie D, Kozakov D, Vajda S. Accelerating and focusing protein-protein docking correlations using multi-dimensional rotational FFT generating functions. Bioinformatics. 2008;24:1865–1873. doi: 10.1093/bioinformatics/btn334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Méndez R, Leplae R, De Maria L, Wodak SJ. Assessment of blind predictions of protein-protein interactions: current status of docking methods. Proteins: Struct. Func. Genet. 2003;52:51–67. doi: 10.1002/prot.10393. [DOI] [PubMed] [Google Scholar]