Abstract
deconSTRUCT webserver offers an interface to a protein database search engine, usable for a general purpose detection of similar protein (sub)structures. Initially, it deconstructs the query structure into its secondary structure elements (SSEs) and reassembles the match to the target by requiring a (tunable) degree of similarity in the direction and sequential order of SSEs. Hierarchical organization and judicious use of the information about protein structure enables deconSTRUCT to achieve the sensitivity and specificity of the established search engines at orders of magnitude increased speed, without tying up irretrievably the substructure information in the form of a hash. In a post-processing step, a match on the level of the backbone atoms is constructed. The results presented to the user consist of the list of the matched SSEs, the transformation matrix for rigid superposition of the structures and several ways of visualization, both downloadable and implemented as a web-browser plug-in. The server is available at http://epsf.bmad.bii.a-star.edu.sg/struct_server.html.
INTRODUCTION
deconSTRUCT is a server aimed at fast culling of a large database of protein structures to find viable candidates for a structural match with the query. The match is reported even if it corresponds only to a substructure of either or both the query and the target. The task is of interest to researchers trying to classify novel structures, model protein structure through assembling pieces of existing structure or transfer the annotation between proteins. The guiding idea in the design of the underlying algorithm has been detection of pairs of protein (sub)structures corresponding to human intuition of the structural match.
Several of deconSTRUCT's features are shared with other published servers, whose explicit purpose is the comparison of a specified structure against a large database of known protein structures: free anonymous access [VASTsearch (1), SSM (2), DaliLite (3), iSARST (4)]; parameter adjustment (iSARST, to certain extent); choice of database against which the search is performed (iSARST, FATCAT); web-browser based [VAST, SSM, FATCAT(5), DaliLite, iSARST, SALAMI (6)] and downloadable (SSM, FATCAT) visualization. To a certain extent, similar information is retrievable from databases of aligned protein structures, such as GANGSTA+ (7), VAST or TOPOFIT (8). Somewhat less related in their designed goal are servers geared strongly toward speed of database search, but operating on the level of full structural domains (9–13), or structural motifs (14). The above list is by no means exhaustive, but, rather, reflects our current understanding of the status of this field of research. For a recent review, see ref. (15).
Of interest to users inclined to a hands-on approach, several methods capable of doing a database-sized search are not (yet) implemented as servers, but can be obtained from their respective websites [SABERTOOTH (16), MAMMOTH (17), TMalign (18), 3dhit (19), the last one also providing a minimalist server, with the list of the top results mailed to the user]. Finally, one should be aware that the methods geared toward a database-sized search are not necessarily the most precise, when it comes to the pairwise alignment of protein structures. Therefore, once the database search is performed, the user (or the implementation) might choose to improve the alignment using one of the slower and more precise methods (20–26).
Ultimately, the reason why all of the above servers coexist, is that they implement different search algorithms, resulting in a somewhat different (and differently ordered) hit list. Aside from the established workhorses in the field, VAST, SSM and DaliLite, of interest, in our possibly biased view, are FATCAT, designed to detect flexible matches in a database search, and iSARST, not only for offering their service using a whopping 80 CPU cluster, but also in that it gives the user a choice of three different alignment improvement engines.
The distinct features of deconSTRUCT are its ability to recognize the match on the level of a substructure, and to report clearly the regions motivating the match, in terms of the implemented visualizations.
METHOD
deconSTRUCT's primary purpose is reduction of the search space by imposing a sequence of requirements that a pair of structures should satisfy in order to constitute a structural match. By its design, it works only for proteins with at least a minimal amount of recognizable secondary structure. The stages of the matching are the following [see Supplementary Data for the precise algorithm used by the search engine, as well as (27) for a more thorough discussion of the underlying ideas]:
Direction matching. The key directions (i.e. the directions determined by helices and strands) in the two structures are required to match. A search in rotational space is performed to establish whether this is the case.
Sequential order checking. The algorithm checks whether the secondary structure elements (SSEs) pointing in the same direction follow the same sequential order in the two structures. The out-of-order SSEs are dropped from further consideration.
Space layout checking. In this final filtering step, the SSEs having the same sequential order in the two proteins, and pointing in the same (within certain tolerance) direction in space are required to occupy the same (again, within tolerance) position in space. If the number of SSEs satisfying all of the conditions so far is non-trivial, the algorithm proceeds to the final steps.
Alignment of matched SSEs on the level of backbone atoms. The backbone alignment is performed as a post-processing step, after the database scan using steps (i) to (iii) is completed. Here, the translation is added and the rotation matrix improved on. The number of top database returns to be handled at this stage can be adjusted by the user.
Alignment extension. To estimate the quality of the overall match in terms of the root-mean-square distance of the paired Cα atoms, the quantity intuitively appealing to many researchers in the field, the transformation is further improved to include as many residues neighboring the matched SSEs as possible.
USER PERSPECTIVE
Input
deconSTRUCT allows several mix-and-match modes of input. A user can specify a PDB (28) identifier of interest, along with the chain, or upload a structure in PDB format, and compare it with one of several non-redundant selections of representative structures (with the sequence identity cutoff ranging between 30% and 100%) or all chains in the PDB. Alternatively, another structure can be specified or uploaded and a one-against-one comparison performed.
The server works with a set of default similarity criteria that can be, optionally, modified by the user through the HTML form on the submission page. In particular, the user can regulate the tightness of the match in SSE direction, the tolerance in the length variation among matched SSEs and the number of hits for which the alignment at backbone level is performed. The defaults are set to work well in an average case, and the interested reader is referred to the Help page of the server for suggestions on tweaking the search toward a more specific target.
Output
The server consists of two main functional branches: database search and post-processing of the results on the pairwise level.
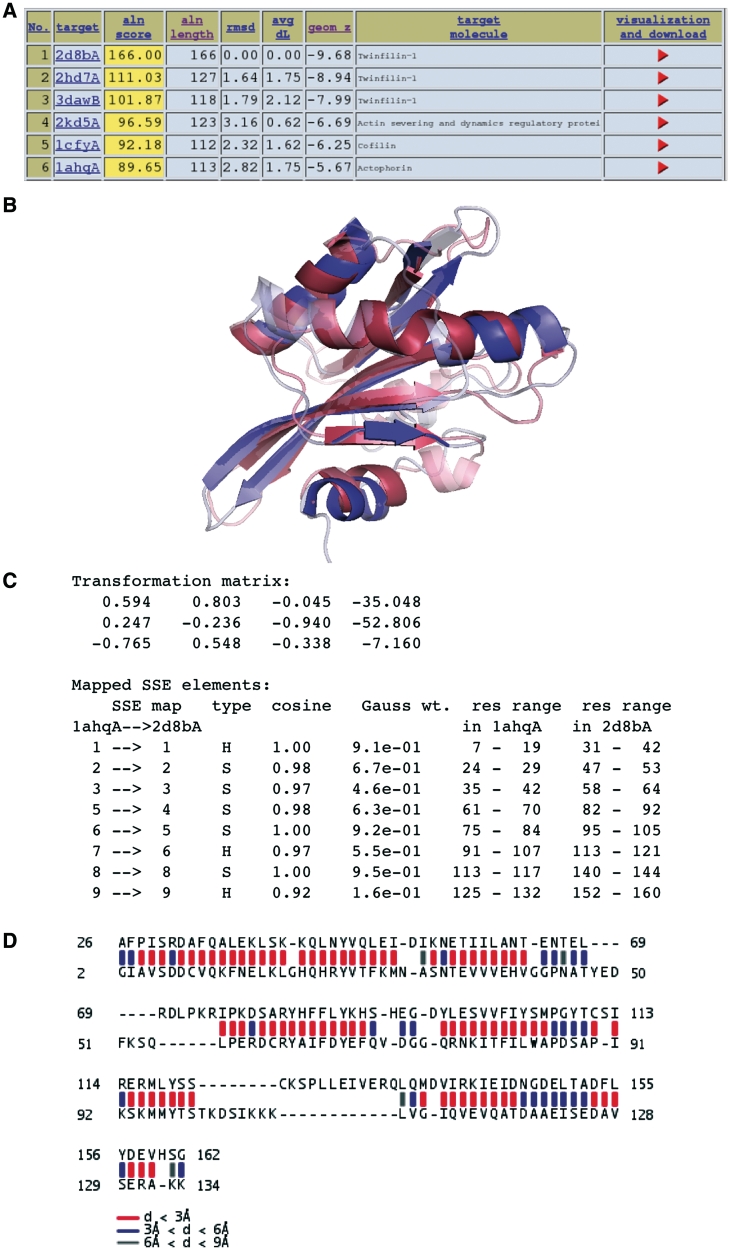
In the case of a one-against-database search, the output consists, in its most elementary form, of a downloadable table of hits. Upon request, this table is emailed to the user. The top hits are also displayed in HTML format, together with the links to the original entries in the PDB database and to the page with more details of the structure comparison for the hit-query pair, Figure 1.
Figure 1.
Result presentation in deconSTRUCT. (A) Results of a database search are presented in form of a table, giving several scores for each hit, and the link (red arrowhead) to the page describing the query-hit match in more detail. (B–D) Query-hit comparison page. (B) The page provides Jmol visualization of the structure superposition, as well as links for the download of Pymol and Chimera sessions using the same visualization scheme: the SSEs motivating the match are represented in solid color, whereas the rest of the two structures is semi-transparent. The visualization using Pymol shown. (C) Furthermore, the page lists the transformation used to produce the coordinate superposition in a typical format: three columns of the rotation matrix, followed by the translation vector column. The transformation applies to the hit structure. Following is the list of mapped elements of secondary structure, including their sequential number, type (strand or helix) cosine of the angle between the matched SSEs, the exponential weight for the cosine (see Supplementary Data, Equation 2) and their range on the respective structure. (D) Finally, the last piece of visualization shows the distance between structurally alignable residues as a colored bar between them, the color indicating the distance range between the corresponding Cαs.
In addition, the server prepares a downloadable Pymol (29) and Chimera (30) sessions. These formats enable the user to download both the superimposed set of coordinates, as well as ready-to-use visualization. For users having different preferences in visualization or post-processing of the results, the superimposed structures can be downloaded in a single PDB file.
In the case of one-to-one comparison, the database search is short-circuited and the pair of structures sent directly to the pairwise analysis stage.
Help
The help page was designed to be as succinct as possible and give the requested information at-a-glance. It is an HTML page with links to its sections included at the top of each search page and from fields in both the input and the output pages.
IMPLEMENTATION AND PERFORMANCE
Implementation
The search engine behind deconSTRUCT is implemented in C. By itself, this implementation is capable of database-against-database comparison and it will be made available in the near future for users wishing to use it in this mode. The front end interface uses Perl/CGI.
Dependencies
Visualization is provided using Jmol (31), and downloadable Pymol (29) and Chimera (30) sessions. For the uploaded structures, the SSEs are assigned using the STRIDE program (32). The representative subsets of the PDB are created using BLASTClust (in our case downloaded from ftp://resources.rcsb.org/sequence/clusters/). The representative sets and the PDB itself are on a monthly update schedule.
Performance
To put the performance of the method behind the server in the context of other currently available methods, we compare the times and sensitivity/specificity tradeoff of deconSTRUCT with CE (20), TopMatch (33) and 3dhit (19), three methods representative of a decade of research in the field. For more extensive testing and comparison with other methods, the reader is referred to ref. (27).
Staying within the scope of this work, we would like to establish the capability and limitations of the method on the particular task of detecting a substructure common to two larger protein structures. Thus, we propose using a test set (available from the server website) consisting of 146 multi-domain chains, with <25% identity between any pair of chains. When deciding on the test set we had to choose a definition of a correct match, a ‘true positive.’ We opted for CATH (34) classification as a guide: a true positive occurs when the query and the target have at least one domain with the same CATH fold-family (‘CAT’ classification). While this definition comes fraught with a certain degree of imprecision, [see for example the discussion in ref. (33) and references therein] it is still a usable tool for comparison as long as all compared methods have to answer the exact same question. In particular, the method showing the highest precision (3dhit in this case) sets the lower bound on what is achievable using a test set and the associated definition of true positives.
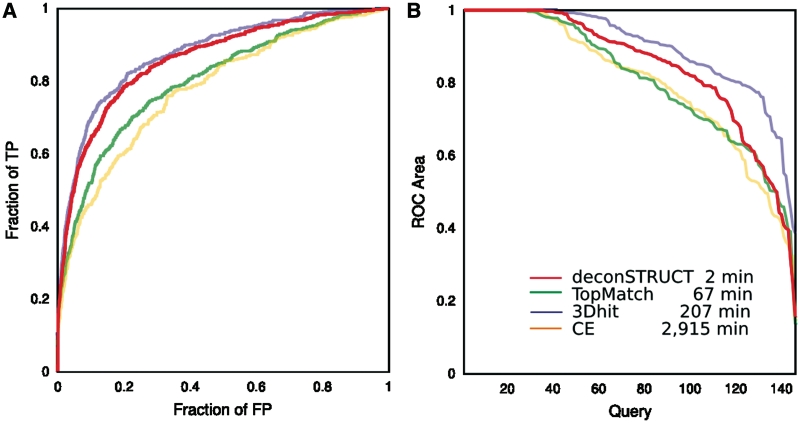
When considering the results in Figure 2 one should keep in mind that CE is a structure alignment program, and if the task was different—specifically, if the task was to find the optimal backbone match given two pieces of structure comparable in size, this and other ‘high resolution’ alignment strategies would probably come closer to the top in performance.
Figure 2.
Performance of the method behind deconSTRUCT, in comparison with other representative methods. The two panels give two representations of data collected in the same computational experiment. The legend corresponds to both panels. For description of the test set, see the main text. The times are CPU times, on a 3 GHz processor. Although the presented graphs use each pairwise comparison once (query-versus-target but not target-versus-query and not query-versus-self), all pairs (including query-versus. self) were used for the timing runs. deconSTRUCT uses pre-processed structure files. Pre-processing of the presented test set takes 4s. If it were it processing two full PDB entries in each pairwise comparison, as the other methods (in the implementation available to us) do, the total deconSTRUCT time would be 15 min. (A) ROC curves. For each method and for every possible pair in the test set, the quality of the structural match is evaluated. The pairs are sorted according to the match score native to each method. The ROC curve shows fraction of true positive versus fraction of false positive as the cutoff in the score value is moved down the sorted pairs list. This graph is a standard way of representing and comparing binary classifiers as their discrimination threshold is varied. (B) ROC area versus query. For each individual query, the area under that query's ROC curve is calculated. For each method, the queries are sorted according to ROC area and the ROC area is plotted as a function of (sorted) query. This plot shows the ability of the method to bring to the top of the list true positives for a given query (irrespective of the values that the scoring function might take for other queries) which is precisely the task of a server, like deconSTRUCT discussed here.
The results in Figure 2 indicate deconSTRUCT's suitability for its proposed task: its speed makes it applicable for searching through large protein structure sets with the performance comparable to the one seen in much more detailed (and therefore slower) methods.
CONCLUSION
deconSTRUCT, the server described in this article, provides access to a method with good tradeoff in sensitivity and speed for a search of structures, or pieces thereof, bearing similarity to the query. Since its speed relies on algorithmic solutions, rather than use of multiple processors or assumed hierarchical organization of protein structures, deconSTRUCT not only offers a new way to perform protein structure comparison and database search, but also comes with multiple possibilities for improvement and growth.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Biomedical Research Council of Agency for Science, Technology and Research Singapore. Funding for open access charge: Biomedical Research Council of Agency for Science, and Research Technology Singapore.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors wish to thank Markus Wiederstein and Shuoyong Shi for their help with TopMatch and ProsMos, respectively. Special thanks to Caleb Khor for his help in setting up, maintaining and improving the server, and Ivica Res for the sequence alignment visualization application. Many thanks, too, to our friends and colleagues in BII, Singapore, for testing and criticizing the server.
REFERENCES
- 1.Madej T, Gibrat J, Bryant S. Threading a database of protein cores. Protein Struct. Funct. Genet. 1995;23:356–369. doi: 10.1002/prot.340230309. [DOI] [PubMed] [Google Scholar]
- 2.Krissinel E, Henrick K. Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr. D. 2004;60:2256–2268. doi: 10.1107/S0907444904026460. [DOI] [PubMed] [Google Scholar]
- 3.Holm L, Kaariainen S, Rosenstrom P, Schenkel A. Searching protein structure databases with DaliLite v. 3. Bioinformatics. 2008;24:2780. doi: 10.1093/bioinformatics/btn507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lo W, Lee C, Lee C, Lyu P. iSARST: an integrated SARST web server for rapid protein structural similarity searches. Nucleic Acids Res. 2009;37:W545–W551. doi: 10.1093/nar/gkp291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ye Y, Godzik A. Flexible structure alignment by chaining aligned fragment pairs allowing twists. Bioinformatics. 2003;19(Suppl. 2):246–255. doi: 10.1093/bioinformatics/btg1086. [DOI] [PubMed] [Google Scholar]
- 6.Margraf T, Schenk G, Torda A. The SALAMI protein structure search server. Nucleic Acids Res. 2009;37(Web Server issue):W480. doi: 10.1093/nar/gkp431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kolbeck B, May P, Schmidt-Goenner T, Steinke T, Knapp E. Connectivity independent protein-structure alignment: a hierarchical approach. BMC Bioinformatics. 2006;7:510. doi: 10.1186/1471-2105-7-510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Leslin C, Abyzov A, Ilyin V. TOPOFIT-DB, a database of protein structural alignments based on the TOPOFIT method. Nucleic Acids Res. 2007;35(Database issue):D317. doi: 10.1093/nar/gkl809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Martin A. The ups and downs of protein topology; rapid comparison of protein structure. Protein Eng. Design Sel. 2000;13:829–837. doi: 10.1093/protein/13.12.829. [DOI] [PubMed] [Google Scholar]
- 10.Roegen P, Fain B. Automatic classification of protein structure by using Gauss integrals. Proc. Natl Acad. Sci. USA. 2003;100:119–124. doi: 10.1073/pnas.2636460100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lisewski A, Lichtarge O. Rapid detection of similarity in protein structure and function through contact metric distances. Nucleic Acids Res. 2006;34:e152. doi: 10.1093/nar/gkl788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Konagurthu A, Stuckey P, Lesk A. Structural search and retrieval using a tableau representation of protein folding patterns. Bioinformatics. 2008;24:645. doi: 10.1093/bioinformatics/btm641. [DOI] [PubMed] [Google Scholar]
- 13.Carpentier M, Brouillet S, Pothier J. YAKUSA: a fast structural database scanning method. Proteins. 2005;61:137–151. doi: 10.1002/prot.20517. [DOI] [PubMed] [Google Scholar]
- 14.Shi S, Chitturi B, Grishin N. ProSMoS server: a pattern-based search using interaction matrix representation of protein structures. Nucleic Acids Res. 2009;37:W526. doi: 10.1093/nar/gkp316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hasegawa H, Holm L. Advances and pitfalls of protein structural alignment. Curr. Opin. Struct. Biol. 2009;19:341–348. doi: 10.1016/j.sbi.2009.04.003. [DOI] [PubMed] [Google Scholar]
- 16.Teichert F, Bastolla U, Porto M. SABERTOOTH: protein structural alignment based on a vectorial structure representation. BMC Bioinformatics. 2007;8:425. doi: 10.1186/1471-2105-8-425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ortiz A, Strauss C, Olmea O. MAMMOTH (matching molecular models obtained from theory): an automated method for model comparison. Protein Sci. 2002;11:2606. doi: 10.1110/ps.0215902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang Y, Skolnick J. TM-align: a protein structure alignment algorithm based on the TM-score. Nucleic Acids Res. 2005;33:2302. doi: 10.1093/nar/gki524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Plewczynski D, Pas J, Von Grotthuss M, Rychlewski L. 3D-Hit, Fast Structural Comparison of Proteins. Appl. Bioinform. 2002;1:223. [PubMed] [Google Scholar]
- 20.Shindyalov I, Bourne P. Protein structure alignment by incremental combinatorial extension (ce) of the optimal path. Protein Eng. 1998;11:739–747. doi: 10.1093/protein/11.9.739. [DOI] [PubMed] [Google Scholar]
- 21.Holm L, Sander C. Dali: a network tool for protein structure comparison. Trends Biochem. Sci. 1995;20:478–480. doi: 10.1016/s0968-0004(00)89105-7. [DOI] [PubMed] [Google Scholar]
- 22.Orengo C, Taylor W. SSAP: sequential structure alignment program for protein structure comparison. Methods Enzymol. 1996;266:617. doi: 10.1016/s0076-6879(96)66038-8. [DOI] [PubMed] [Google Scholar]
- 23.Gerstein M, Levitt M. Comprehensive assessment of automatic structural alignment against a manual standard, the SCOP classification of proteins. Protein Sci. 1998;7:445. doi: 10.1002/pro.5560070226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhu J, Weng Z. FAST: a novel protein structure alignment algorithm. Proteins Struct. Funct. Bioinform. 2005;58:618–627. doi: 10.1002/prot.20331. [DOI] [PubMed] [Google Scholar]
- 25.Mosca R, Brannetti B, Schneider T. Alignment of protein structures in the presence of domain motions. BMC Bioinformatics. 2008;9:352. doi: 10.1186/1471-2105-9-352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sippl M. On distance and similarity in fold space. Bioinformatics. 2008;24:872. doi: 10.1093/bioinformatics/btn040. [DOI] [PubMed] [Google Scholar]
- 27.Zhang ZH, Lee HK, Mihalek I. Reduced representation of protein structure: implications on efficiency and scope of detection of structural similarity. BMC Bioinformatics. 2010;11:155. doi: 10.1186/1471-2105-11-155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Berman H, Westbrook J, Feng Z, Gilliland G, Bhat T, Weissig H, Shindyalov I, Bourne P. The protein data bank. Nucletc Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.DeLano W. The PyMOL Molecular Graphics System. 2002 www.pymol.org. [Google Scholar]
- 30.Pettersen E, Goddard T, Huang C, Couch G, Greenblatt D, Meng E, Ferrin T. UCSF Chimera visualization system for exploratory research and analysis. J. Comp. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 31. Jmol: an open-source Java viewer for chemical structures in 3D. www.jmol.org. [Google Scholar]
- 32.Frishman D, Argos P. Knowledge-based protein secondary structure assignment. Proteins Struct. Funct. Genet. 1995;23:566–579. doi: 10.1002/prot.340230412. [DOI] [PubMed] [Google Scholar]
- 33.Sippl M, Wiederstein M. A note on difficult structure alignment problems. Bioinformatics. 2008;24:426. doi: 10.1093/bioinformatics/btm622. [DOI] [PubMed] [Google Scholar]
- 34.Orengo C, Michie A, Jones S, Jones D, Swindells M, Thornton J. CATH – a hierarchic classification of protein domain structures. Structure. 1997;5:1093–1108. doi: 10.1016/s0969-2126(97)00260-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.