Abstract
e-LEA3D web server integrates three complementary tools to perform computer-aided drug design based on molecular fragments. In drug discovery projects, there is a considerable interest in identifying novel and diverse molecular scaffolds to enhance chances of success. The de novo drug design tool is used to invent new ligands to optimize a user-specified scoring function. The composite scoring function includes both structure- and ligand-based evaluations. The de novo approach is an alternative to a blind virtual screening of large compound collections. A heuristic based on a genetic algorithm rapidly finds which fragments or combination of fragments fit a QSAR model or the binding site of a protein. While the approach is ideally suited for scaffold-hopping, this module also allows a scan for possible substituents to a user-specified scaffold. The second tool offers a traditional virtual screening and filtering of an uploaded library of compounds. The third module addresses the combinatorial library design that is based on a user-drawn scaffold and reactants coming, for example, from a chemical supplier. The e-LEA3D server is available at: http://bioinfo.ipmc.cnrs.fr/lea.html.
INTRODUCTION
Computer-aided drug design methods contribute to the early stage of the drug discovery process to identify new bioactive molecules. Computational methods comprise virtual screening of available chemical databases and de novo drug design. The two approaches aim to select a set of molecules that are predicted to exhibit a biological activity on a given target. The estimation of the activity, usually by the mean of a score, is addressed by various ligand- and structure-based methods depending on the data on known ligands and the availability of the 3D experimental structure of the target.
Contrary to virtual screenings used to mine in-house and commercial collections, de novo drug design can create molecules that do not exist in known compound databases. De novo design methods are automated computational procedures that build molecules by using atoms or fragments with the aim that the resulting molecular structures would fit specified property constraints. They allow the exploration of the theoretically available chemical space, a space larger than that can be enumerated by synthesis (1,2) or even by computer. For example, Blum and Reymond propose a list of small organic molecules containing up to 13 atoms of C, N, O, S and Cl. The public database GDB-13 contains 970 million of druglike molecules (3). Nevertheless, the virtual screening of millions of compounds using docking software, for example, is still time consuming even with high-throughput methods. On the contrary, alternative approaches as de novo drug design use search strategies to efficiently explore the chemical space without a fully enumeration. Particle Swarm Optimization (4) or Evolutionary Algorithms (5), analogous to Genetic Algorithms (6–9), are examples of heuristics employed for this purpose. The main downside of this strategy is that it can generate compounds that are promising but difficult to synthesize (10). To overcome this defect, the designed compounds can alternatively be used as a query in a structural similarity search in commercial collections [e.g. ZINC, http://zinc.docking.org/choose.shtml (11)]. Thus, it is possible to select a small- and focused-library of available analogs that can be further evaluated by a virtual screening step and eventually tested experimentally (12). Likewise, de novo drug design approach has the property to suggest new scaffold candidates. In this case, synthetic chemistry experts may help in defining a synthetic route and in selecting possible reactants to create a combinatorial library (13). For example, van Hoorn and Bell designed a Bayesian Idea Generator to identify the likely chemistry protocols associated with a given compound (14).
The synthesis of combinatorial libraries is another important strategy in drug discovery. This may be achieved by a traditional chemistry synthesis but also, in some particular cases, by a target-guided synthesis (TGS) also called in situ click chemistry (15). TGS lets the protein-binding site to select fittest reagents to form highly potent ligands. For example, Kolb and co-workers discovered new inhibitors for the acetylcholinesterase (AChE) and the carbonic anhydrase II from a (1,3)-dipolar cycloaddition between azides and acetylenes reactants (16–18). This type of approach uses multireagent mixtures whose complexity in terms of components must be limited to avoid aggregation and product identification problems. A virtual screening may help to prioritize the most promising ones when hundreds of reagents are commercially available.
Historically, computational tools have been developed inside companies by modeling teams or by modeling software companies (19,20). Various innovative tools have also been developed by academics but their acquisition by software companies often limited their use for free by this community. As a result, the number of ‘free of charge’ databases and softwares dedicated to cheminformatic is scarce. The CCL computational chemistry provides downloadable software packages, for example, the de novo drug design program NEWLEAD (21) (http://www.ccl.net/cca/software/MAC/newlead_linux_and_mac_osx/). In the same manner, there is a relative few number of web resources dedicated to small molecules compared to other categories (http://bioinformatics.ca/links_directory/). For example, a tremendous number of bioinformatic tools have been created and provided to the entire scientific community to respond to an increasing need in terms of sequence analyses and structure predictions. Few years ago, Villoutreix et al. cataloged a list of resources ranging from homology modeling to protein docking and virtual screening (http://www.vls3d.com/links.html) (22). However, only a minority of chemistry-oriented tools is true on line facility and thus the others are not easily practicable for non-expert scientists. Among web servers, some are dedicated to the virtual screening by docking: DOCK Blaster (http://blaster.docking.org/start.shtml) (23), TarFisDock (http://www.dddc.ac.cn/tarfisdock/) (24) or the SCFBIO server (http://www.scfbio-iitd.res.in/bioinformatics/drugdesign.htm), but they only screen existing, commercial or uploaded, libraries of molecules. This prompts us to develop a cheminformatic web server also able to create and build small molecules.
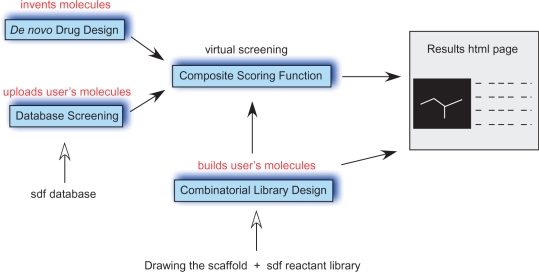
Herein, we present e-LEA3D, the first web server to perform computer-aided de novo drug design, to build focused combinatorial libraries of molecules and to perform virtual screenings. The interplay between these approaches is described in Figure 1. The de novo drug design tool creates new molecules by using a genetic algorithm to evolve a population of molecules which are gradually improved by competing for the ‘survival of the fittest’ (25). Here, a molecule results from the association of various 3D fragments extracted from known bioactives compounds. The second tool assists the user in the design of a combinatorial library where a central core is connected to reactants coming, for example, from a supplier. The reactants are modified according to the user specifications in order to extract the substituent part only. The generated virtual library can be automatically evaluated by using the same scoring function as the one used in the de novo drug design step. It aims to prioritize the reactants.
Figure 1.
Interplay between the three e-LEA3D modules.
CONCEPTS AND METHODS
De novo drug design
The de novo drug design module is based on a new version of the LEA3D engine (25). This program creates new molecules either from scratch or based on a user-defined scaffold on which substituents have to be optimized. The procedure is a fragment-based approach that uses a genetic algorithm to optimize the combination of fragments. The algorithm dynamically evolves a population of molecules through their modifications by mutation and crossover operators. The reproduction is fitness-proportionate to favor best candidate molecules for breeding. At each iteration, a new population of molecules replaces the parents except the best one to comply with the elitism strategy. This prevents the best solution from being lost and allows a relative high percentage of mutations (70%) compared to the crossover one (30%). The mutation operators bring diversity by integrating new fragments in molecules while the crossover operator recombines fragments already present in the current population. During a crossover, two parent molecules interchange a portion of their structures. To mutate molecules, four operators have been created: the suppression of one fragment, the addition or the replacement of one fragment by a new one coming from the fragment database and the permutation or scrambling of fragments of the parent structure. A more detailed description can be found in a previous publication (25). The current version of LEA3D differs from the published one in its way to store the fragments, to represent the molecule and to display the results. Also, it uses different docking program: PLANTS (26) instead of FlexX (27) but the genetic algorithm and its parameters are unchanged.
The current building blocks, rings and acyclic fragments, have been extracted from approved and investigational drugs identified by a USAN (Comprehensive Medicinal Chemistry, Symyx Solutions, Inc.). The fragment database contains 5283 building blocks; only 13% of the disconnected fragments were unique. Each fragment possesses one or more ‘X’ dummy atoms that memorize the original substitution pattern of the fragment as well as the stereochemistry. If a fragment contains several substitution sites, one of them is randomly selected to be connected. Eventually, the 3D conformation of the created molecule is generated by the program Frog (28).
Then, the fitness of each molecule is evaluated via a function which takes as input the molecular structure and returns a numeric score. The evaluation can integrate a selected number of molecular properties and/or a protein–ligand docking score calculated by the program PLANTS (26). The user can define the relative importance of each individual property by assigning it a weight. Ligand-based properties include descriptors commonly used to define the boundaries of the lead-like and drug-like chemical space. The range limits are given for some of them along with a publication reference (molecular weight, Logp, number of atoms, number of h-donors and h-acceptors, polar solvent accessible surface area, molecular refractivity, moment of inertia, rotatable bonds, number of rings and number of aromatic rings). Additional molecular properties are the search of chemical functions, the definition of a pharmacophore and the ligand similarity measure based on a molecular fingerprint. The fingerprint consists of a vector of 120 cells, each cell indicating the presence or the absence of the associated atom type calculated following the definition of Ghose (29,30).
In general, LEA3D produces different solutions at each run since it uses a heuristic and randomly selects fragments and their substitution sites, excepting when it must deduce a chemical structure with descriptor values identical to those of a given target compound. For example, to deduce the structure of the protonated form of aspirin, one uses the following properties: a molecular weight equals to 180, a number of heavy atoms equals to 13, a number of aromatic rings equals to 1 and the presence of the chemical functions ester and acid. In such case, different runs converge toward analogs (score ≤100%) or find the target solution (score=100%).
Finally, e-LEA3D can be set to maintain the presence of one particular fragment or scaffold through the evolutionary process. Subsequently, molecules are evolved by adding one or more substituents to this scaffold at chosen positions. This strategy is advantageous when the docking program, for example FlexX (27), is able to superimpose the conserved building block with a structure whose coordinates in the binding site are known by X-ray crystallography (25); the docking procedure is accelerated.
Combinatorial library builder
Since the 1990s, the experimental combinatorial chemistry and the computational design form a partnership to focus the synthesized collections toward a specified family target and to optimize the ADME characteristics (31). A key feature in e-LEA3D is the possibility to define a specific scaffold as the central core of a combinatorial library. This has been developed to deal with simple cases of library design where a final scaffold is common to all products. Note that this module does not allow reaction-based specifications such as a rearrangement of the central core or of the reactants. To fully exploit this module, a minimal expertise in chemistry is required. To design a library, the user simply draws a scaffold and uploads the reactant file. The procedure is iterative and several substituents may be added to the scaffold. For this purpose, reactants coming, for example, from a chemical supplier are modified according to the user specifications in order to extract the substituent part only. Then, substituents are connected to the scaffold at the selected position. Once created, the library can be evaluated by a composite scoring function (described in the previous section) and thereby, the most promising reagents are prioritized. A tutorial to build an azide/acetylene library is present on our web server. This is a reaction well-suited for the in situ click chemistry (http://bioinfo.ipmc.cnrs.fr/images/tutorial_combinatorial_library.pdf).
WEB SERVER
In both modules, a two steps procedure is required to launch the task. First, the user uploads the input data whose integrity is checked by the server and then, the user is invited to complete the request. Once the run is submitted, a transition page gives a link to the result web page. We encourage the bookmarking of the link. Time-consuming tasks such as the structure-based de novo drug design (∼6 h) and the virtual screening of more than 50 molecules are scheduled by the queuing system Torque. Other tasks may also take long times in case where there are several users employing the web service but usually, they takes few seconds or minutes. The results are stored on the server for 20 days. Tools such as the ACD structure drawing Applet (v.1.30), the Chemis3D (v2.89a) and JMol (v11.6.13) applets allow sketching small molecules and visualizing them at the different steps (input and output).
De novo drug design server
Input
In the front-page, the user is invited to create its composite scoring function by selecting at least one property. Each selected ligand-based property is defined by a minimal and/or maximal value and a weight. A guideline at the bottom of the page explains how to set an exact value or a range of values. Most of the present molecular properties have been used and referenced in the literature to define the limits of an appropriate chemical space. In such cases, default values have been set. Additionally, three other ligand-based properties complement the list: the identification of mandatory chemical characteristics such as chemical functions, the identification of a pharmacophore and a similarity measure based on a fingerprint. In the last case, the similarity between a candidate and a reference molecule is calculated by comparing their atom type compositions calculated following the definition of Ghose and Crippen (GC descriptors) (30).
If the structure-based property is integrated in the function score, it is necessary to upload the protein structure in mol2 format [input for PLANTS (26)]. This format is correctly generated if the protein is protonated. As crystallographic files from the Protein Data Bank do not contain hydrogen atoms, it is necessary to convert the PDB input file into a protonated pdb file by using the PDB2PQR web server (see the help section at the bottom of the front-page) (32). The generated pdb file is suitable to be uploaded by e-LEA3D in order to be converted into a mol2 file format. Finally, it is required to define the binding site by indicating either the name, the number and the chain name of one residue, for example the ligand, present in the pocket or the coordinates of the center of it. The binding site is centered on the last and includes residues whose at least one atom belongs to the sphere of the chosen radius (the default one is 10 Å).
Once the checking step completes, a web page summarizes the scoring function content. At this stage, two additional parameters can be set to generate conformers and to perform the ionization of carboxylate, phosphate, amidinium and guanidinium groups. The default settings activate the generation of one conformer and accept the ionization step. Then, the user can either screen a library of molecules (‘Database screening’ button on the right) or start the ‘De novo Drug Design’ program on the left. The ‘Database screening’ option needs to upload a sdf file of the molecules to screen. An option enables to filter out unfitted molecules (i.e. having a score <100%). This option is omitted when the docking is a part of the scoring function. In the ‘De novo Drug Design’ program, most of the parameters are already defined but it is possible to modify the population size and the number of generations. Note that changing these parameters is not recommended as it can drastically increase the time-cost of the drug design process. An advanced option allows selecting a mandatory scaffold/fragment in sdf format that must be present in each generated molecule. This option is useful when the objective is to search new substituents or to growth a fragment known to bind the protein target.
The computational procedure filters out candidate molecules that have not 80% of the molecular properties when structure- and molecular-based properties are selected. Indeed, the docking step and the 3D conformation generation are the most time consuming tasks. Therefore, it is recommended to associate the docking function with an upper limit for the molecular weight (MW ≤500 for example).
Output
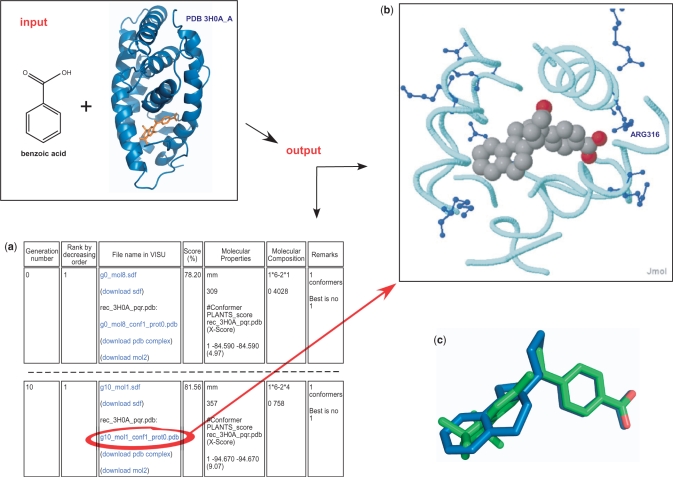
The Figure 2 presents an example of the output page resulting from the automated de novo drug design of ligands targeting the retinoic acid receptor RXRα and having a benzoic acid as a mandatory fragment. The right panel of the web page is updated every minute to follow the design over the generations. Each row summarizes the features of the best candidate of the generation: the generation number, the rank, the score in percentage (the maximum is 100%), the molecular properties of the selected conformer, the fragment composition and the number of the best conformer. The left panel displays the molecule in 3D alone or in complex with a binding site if the scoring function includes a docking score. A click on the molecule name at the right panel executes the display at the left window. Molecules and complexes are also downloadable in sdf, mol2 and pdb file format, respectively. If several conformers have been generated for one molecule, the sdf file contains the coordinates of the best one whereas the mol2 file contains its docking pose conformation. There is also a link at the top of the right page to download the concatenated sdf file of the best candidates. Once the run completes, two links give access to the convergence plot of the genetic algorithm for the mean score and for the best score.
Figure 2.
Input and Output contents of the de novo drug design module. Inputs are the structure of the RXRα nuclear receptor and the building block ‘benzoic acid’, a substructure of the approved drug bexarotene present in the crystallized structure (residue name 9RA; the picture has been produced using PyMOL). The composite scoring function includes a docking by the program PLANTS in the RXRα binding site and a calculation of the molecular weight that must be ≤469. Outputs are: (a) the follow-up of the fittest molecule of each generation. Only the first and the last generation are presented. The score in percentage is displayed (81.56% for the best candidate) as well as its composition: the PLANTS score (–94.67) and the molecular weight (357). A second score in parenthesis, the Xscore (37), evaluates the docking pose from PLANTS (9.07). The molecular composition and the conformer number are given. The best candidate is a combination of the building block ‘benzoic acid’ (lego 0) with the lego number 758. The first building block (lego 0) is connected by its atom number 6 (coded by 1*6) with the second building block (lego 758) by its atom number 4 (2*4). (b) A click on the file name in blue displays the molecule in the java applet Jmol. The backbone of the active site is depicted in cyan and the side chains of basic residues are colored in blue. The side chain of the ARG316 is involved in an ionic bond with the carboxylate of the ligand whose carbons are colored in grey and oxygens in red. (c) The superimposition of the de novo candidate in blue on the co-crystallized drug bexarotene in green. The benzoic acid groups are perfectly superimposed.
Combinatorial library builder server
Input
The user is guided step by step to define the building blocks of the combinatorial library. First, the final scaffold is drawn by using the ACD structure drawing Applet. Then, it must be converted into a text file by the button ‘Click to Convert’ and submitted to a 3D conversion. Note that the positions of the future substituents are linked to a hydrogen atom. The result web page displays the sketch of the central core with its formula, molecular weight and a link to download the sdf file. The Chemis3D applet displays the fragment and the numbering of the atoms and thus it enables to identify the number of the heavy atom that will be substituted. A field is allocated to store this number. Then, the user uploads the reagent file in sdf format. The reagent molecules must have a common functional group such as an amine, a thiol, a bromine atom … The next step aims to define where to break a single bond to extract the substituent part. Two cases may occur. The first case allows identifying the atom type to be replaced by the final scaffold. Additional information about the neighbor atoms may help to select the right site and the ‘keep’ comment indicates which fragment of the two disconnected parts is the substituent. The second case has been created to deal with reagents whose atom to be replaced may be ambiguous like a hydrogen atom for example. To localize the appropriate hydrogen, the user must define an unambiguous neighbor, for example, a sulfur atom connected to the hydrogen to replace. This neighbor atom can be further described by its bonded neighbors and the comment ‘keep’ is recommended to flag the desirable part. For each case, an example is displayed at the right side of the field. Finally, a checkbox offers the option to generate a conformer for each combined molecule but this option must not be selected if the user plans to add another substituent because the order of the atoms in the sdf file is scrambled by the 3D generator program.
Output
The output web page presents four parts from (A) to (C.2). The first section summarizes the reagent modifications by visualizing the first reagent of the upload file and the first modified reagent. The ‘X’ atom in pink in the Chemis3D display is replaced by the final scaffold. The accepted reagent file is downloadable in sdf format. The second section (B) provides a link to the result web page similar to that one generated by the de novo drug design server. Each combined molecule is visualized by the Jmol applet. In the section (C), the combinatorial library can be either evaluated by a scoring function or modified by adding another reagent. In the (C.1) section, a link heads to the front-page of the ‘de novo drug design or screen’ module. Once the scoring function is built, the ‘Database screening’ module automatically gets the combinatorial library as the database to screen. The result web page presents the same features as in (B). In addition, it recapitulates the sdf datablock of the reagents in the molecular properties column when it exists in the original file. This is useful to identify the reagents by their supplier identifiers. In the (C.2) section, the user can iterate the addition of another reagent. It produces a result page with the same features as the present one.
CONCLUSIONS
We presented e-LEA3D, a server that aims to invent ideas of ligands (scaffold-hopping), to screen/dock user’s molecules into the structure of the target protein and to assist scientists in the optimization of hits by the creation of a focused combinatorial library of molecules. The discovery of new potent ligands remains crucial for a large spectrum of applications and topics as pharmacological tools for in vitro and in vivo physiophatologic studies. This server complements other services dedicated to small molecules and which are presented in a pipeline manner: PharmaGist for a pharmacophore detection based on aligned ligands (33), Superimposé for a 3D similarity search of known ligands (34), wwLigCSRre for a similarity search in commercial databases (35) and FAF-Drugs for the ADME/tox filtering of candidates (36). In the near future, we plan an upgrade of the ‘Database screening’ module where we will offer the selection of several focused databases to screen approved drugs and commercial subdrugs of drugs suitable, for example, to start a fragment-based drug discovery program.
FUNDING
Funding for open access charge: French National Research Agency (Grant FRAGSCREEN ANR-07-JC-JC-0046-01).
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors thank Thomas Exner for the permission to use PLANTS on the server. The authors also thank the developers of free and/or open source software: Frog, Jmol, Chemis3D, mol2ps, ACDLABS for the structure drawing applet and OpenBabel.
REFERENCES
- 1.Lipinski C, Hopkins A. Navigating chemical space for biology and medicine. Nature. 2004;432:855–861. doi: 10.1038/nature03193. [DOI] [PubMed] [Google Scholar]
- 2.Triggle DJ. The chemist as astronaut: searching for biologically useful space in the chemical universe. Biochem. Pharmacol. 2009;78:217–223. doi: 10.1016/j.bcp.2009.02.015. [DOI] [PubMed] [Google Scholar]
- 3.Blum LC, Reymond JL. 970 million druglike small molecules for virtual screening in the chemical universe database GDB-13. J. Am. Chem. Soc. 2009;131:8732–8733. doi: 10.1021/ja902302h. [DOI] [PubMed] [Google Scholar]
- 4.Hartenfeller M, Proschak E, Schuller A, Schneider G. Concept of combinatorial de novo design of drug-like molecules by particle swarm optimization. Chem. Biol. Drug Des. 2008;72:16–26. doi: 10.1111/j.1747-0285.2008.00672.x. [DOI] [PubMed] [Google Scholar]
- 5.Masek BB, Shen L, Smith KM, Pearlman RS. Sharing chemical information without sharing chemical structure. J. Chem. Inf. Model. 2008;48:256–261. doi: 10.1021/ci600383v. [DOI] [PubMed] [Google Scholar]
- 6.Wang R, Gao Y, Lai L. LigBuilder: a multi-purpose program for Structure-based drug design. J. Mol. Model. 2000;6:498–516. [Google Scholar]
- 7.Glen RC, Payne AW. A genetic algorithm for the automated generation of molecules within constraints. J. Comput. Aided Mol. Des. 1995;9:181–202. doi: 10.1007/BF00124408. [DOI] [PubMed] [Google Scholar]
- 8.Pegg SC, Haresco JJ, Kuntz ID. A genetic algorithm for structure-based de novo design. J. Comput. Aided Mol. Des. 2001;15:911–933. doi: 10.1023/a:1014389729000. [DOI] [PubMed] [Google Scholar]
- 9.Clark DE, Frenkel D, Levy SA, Li J, Murray CW, Robson B, Waszkowycz B, Westhead DR. PRO-LIGAND: an approach to de novo molecular design. 1. Application to the design of organic molecules. J. Comput. Aided Mol. Des. 1995;9:13–32. doi: 10.1007/BF00117275. [DOI] [PubMed] [Google Scholar]
- 10.Stahl M, Todorov NP, James T, Mauser H, Boehm HJ, Dean PM. A validation study on the practical use of automated de novo design. J. Comput. Aided Mol. Des. 2002;16:459–478. doi: 10.1023/a:1021242018286. [DOI] [PubMed] [Google Scholar]
- 11.Irwin JJ, Shoichet BK. ZINC–a free database of commercially available compounds for virtual screening. J. Chem. Inf. Model. 2005;45:177–182. doi: 10.1021/ci049714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Honma T. Recent advances in de novo design strategy for practical lead identification. Med. Res. Rev. 2003;23:606–632. doi: 10.1002/med.10046. [DOI] [PubMed] [Google Scholar]
- 13.Leach AR, Bryce RA, Robinson AJ. Synergy between combinatorial chemistry and de novo design. J. Mol. Graph Model. 2000;18:358–367, 526. doi: 10.1016/s1093-3263(00)00062-0. [DOI] [PubMed] [Google Scholar]
- 14.van Hoorn WP, Bell AS. Searching chemical space with the Bayesian Idea Generator. J. Chem. Inf. Model. 2009;49:2211–2220. doi: 10.1021/ci900072g. [DOI] [PubMed] [Google Scholar]
- 15.Schmidt MF, Rademann J. Dynamic template-assisted strategies in fragment-based drug discovery. Trends Biotechnol. 2009;27:512–521. doi: 10.1016/j.tibtech.2009.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Manetsch R, Krasinski A, Radic Z, Raushel J, Taylor P, Sharpless KB, Kolb HC. In situ click chemistry: enzyme inhibitors made to their own specifications. J. Am. Chem. Soc. 2004;126:12809–12818. doi: 10.1021/ja046382g. [DOI] [PubMed] [Google Scholar]
- 17.Mocharla VP, Colasson B, Lee LV, Roper S, Sharpless KB, Wong CH, Kolb HC. In situ click chemistry: enzyme-generated inhibitors of carbonic anhydrase II. Angew. Chem. Int. Ed. Engl. 2004;44:116–120. doi: 10.1002/anie.200461580. [DOI] [PubMed] [Google Scholar]
- 18.Bourne Y, Kolb HC, Radic Z, Sharpless KB, Taylor P, Marchot P. Freeze-frame inhibitor captures acetylcholinesterase in a unique conformation. Proc. Natl Acad. Sci. USA. 2004;101:1449–1454. doi: 10.1073/pnas.0308206100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Richon AB. Current status and future direction of the molecular modeling industry. Drug Discov. Today. 2008;13:665–669. doi: 10.1016/j.drudis.2008.04.008. [DOI] [PubMed] [Google Scholar]
- 20.Richon AB. An early history of the molecular modeling industry. Drug Discov. Today. 2008;13:659–664. doi: 10.1016/j.drudis.2008.03.012. [DOI] [PubMed] [Google Scholar]
- 21.Tschinke V, Cohen NC. The NEWLEAD program: a new method for the design of candidate structures from pharmacophoric hypotheses. J. Med. Chem. 1993;36:3863–3870. doi: 10.1021/jm00076a016. [DOI] [PubMed] [Google Scholar]
- 22.Villoutreix BO, Renault N, Lagorce D, Sperandio O, Montes M, Miteva MA. Free resources to assist structure-based virtual ligand screening experiments. Curr. Protein Pept. Sci. 2007;8:381–411. doi: 10.2174/138920307781369391. [DOI] [PubMed] [Google Scholar]
- 23.Irwin JJ, Shoichet BK, Mysinger MM, Huang N, Colizzi F, Wassam P, Cao Y. Automated docking screens: a feasibility study. J. Med. Chem. 2009;52:5712–5720. doi: 10.1021/jm9006966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li H, Gao Z, Kang L, Zhang H, Yang K, Yu K, Luo X, Zhu W, Chen K, Shen J, et al. TarFisDock: a web server for identifying drug targets with docking approach. Nucleic Acids Res. 2006;34:W219–W224. doi: 10.1093/nar/gkl114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Douguet D, Munier-Lehmann H, Labesse G, Pochet S. LEA3D: a computer-aided Ligand design for structure-based drug design. J. Med. Chem. 2005;48:2457–2468. doi: 10.1021/jm0492296. [DOI] [PubMed] [Google Scholar]
- 26.Korb O, Stutzle T, Exner TE. Empirical scoring functions for advanced protein-ligand docking with PLANTS. J. Chem. Inf. Model. 2009;49:84–96. doi: 10.1021/ci800298z. [DOI] [PubMed] [Google Scholar]
- 27.Rarey M, Kramer B, Lengauer T, Klebe G. A fast flexible docking method using an incremental construction algorithm. J. Mol. Biol. 1996;261:470–489. doi: 10.1006/jmbi.1996.0477. [DOI] [PubMed] [Google Scholar]
- 28.Leite TB, Gomes D, Miteva MA, Chomilier J, Villoutreix BO, Tuffery P. Frog: a FRee Online druG 3D conformation generator. Nucleic Acids Res. 2007;35:W568–W572. doi: 10.1093/nar/gkm289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Roche O, Schneider P, Zuegge J, Guba W, Kansy M, Alanine A, Bleicher K, Danel F, Gutknecht EM, Rogers-Evans M, et al. Development of a virtual screening method for identification of “frequent hitters” in compound libraries. J. Med. Chem. 2002;45:137–142. doi: 10.1021/jm010934d. [DOI] [PubMed] [Google Scholar]
- 30.Viswanadhan VN, Ghose AK, Revankar GR, Robins RK. Atomic physicochemical parameters for three dimensional structure directed quantitative structure-activity relationships. J. Chem. Inf. Comput. Sci. 1989;29:163–172. doi: 10.1021/ci00053a005. [DOI] [PubMed] [Google Scholar]
- 31.Moos WH, Hurt CR, Morales GA. Combinatorial chemistry: oh what a decade or two can do. Mol. Divers. 2009;13:241–245. doi: 10.1007/s11030-009-9127-y. [DOI] [PubMed] [Google Scholar]
- 32.Dolinsky TJ, Nielsen JE, McCammon JA, Baker NA. PDB2PQR: an automated pipeline for the setup of Poisson-Boltzmann electrostatics calculations. Nucleic Acids Res. 2004;32:W665–W667. doi: 10.1093/nar/gkh381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schneidman-Duhovny D, Dror O, Inbar Y, Nussinov R, Wolfson HJ. PharmaGist: a webserver for ligand-based pharmacophore detection. Nucleic Acids Res. 2008;36:W223–W228. doi: 10.1093/nar/gkn187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bauer RA, Bourne PE, Formella A, Frommel C, Gille C, Goede A, Guerler A, Hoppe A, Knapp EW, Poschel T, et al. Superimpose: a 3D structural superposition server. Nucleic Acids Res. 2008;36:W47–W54. doi: 10.1093/nar/gkn285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sperandio O, Petitjean M, Tuffery P. wwLigCSRre: a 3D ligand-based server for hit identification and optimization. Nucleic Acids Res. 2009;37:W504–W509. doi: 10.1093/nar/gkp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Miteva MA, Violas S, Montes M, Gomez D, Tuffery P, Villoutreix BO. FAF-Drugs: free ADME/tox filtering of compound collections. Nucleic Acids Res. 2006;34:W738–W744. doi: 10.1093/nar/gkl065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang R, Lai L, Wang S. Further development and validation of empirical scoring functions for structure-based binding affinity prediction. J. Comput. Aided Mol. Des. 2002;16:11–26. doi: 10.1023/a:1016357811882. [DOI] [PubMed] [Google Scholar]