Abstract
In silico drug target identification, which includes many distinct algorithms for finding disease genes and proteins, is the first step in the drug discovery pipeline. When the 3D structures of the targets are available, the problem of target identification is usually converted to finding the best interaction mode between the potential target candidates and small molecule probes. Pharmacophore, which is the spatial arrangement of features essential for a molecule to interact with a specific target receptor, is an alternative method for achieving this goal apart from molecular docking method. PharmMapper server is a freely accessed web server designed to identify potential target candidates for the given small molecules (drugs, natural products or other newly discovered compounds with unidentified binding targets) using pharmacophore mapping approach. PharmMapper hosts a large, in-house repertoire of pharmacophore database (namely PharmTargetDB) annotated from all the targets information in TargetBank, BindingDB, DrugBank and potential drug target database, including over 7000 receptor-based pharmacophore models (covering over 1500 drug targets information). PharmMapper automatically finds the best mapping poses of the query molecule against all the pharmacophore models in PharmTargetDB and lists the top N best-fitted hits with appropriate target annotations, as well as respective molecule’s aligned poses are presented. Benefited from the highly efficient and robust triangle hashing mapping method, PharmMapper bears high throughput ability and only costs 1 h averagely to screen the whole PharmTargetDB. The protocol was successful in finding the proper targets among the top 300 pharmacophore candidates in the retrospective benchmarking test of tamoxifen. PharmMapper is available at http://59.78.96.61/pharmmapper.
INTRODUCTION
Recent advances in genomics have triggered a shift in drug discovery from the paradigm of focusing on strong single-target interaction to more global and comparative analysis of multi-targets network (1–3). In this context, it has become an urgent need to develop fast, robust and efficient methods to identify and validate new druggable targets and, concomitantly, to map the ligand-target profiling space globally. A proteomic approach in identifying potential binding proteins for a given small molecule involves comparison of the protein expression profiles for a given cell or tissue in the presence or absence of the given molecule. This method has not proved very successful in target discovery because it is laborious and time-consuming (4). Within this new scenario, in silico target profiling methods are emerging as efficient alternatives to the currently unaffordable high-throughput in vitro target profiling of compounds as well as to find new therapeutic indications for old drugs, an activity often referred to as drug repurposing (3,5–7). On the other hand, chemogenomics approach has emerged as a new discipline in target prediction via data mining in target-annotated databases (8–15). However, the success of chemogenomics depends on the availability of bioactivity data for the targets and their associated ligands. For new ligands, such data are either approximate or unavailable in lack of corresponding target information. Moreover, the adverse drug reaction may involve targets that are not well-characterized (16). Recently, we have developed an in silico target prediction method for a given small molecule by ‘probing’ the potential ligand binding sites stored in potential drug target database (PDTD) via ligand–protein reverse docking strategy (17,18). As a complementary modeling method to 3D structures at atomic level, pharmacophore is the spatial arrangement of features that enables a molecule to interact with a target receptor in a specific binding mode. Recent developments as well as applications of pharmacophore model derived from protein-ligand 3D complex structures (19,20) have triggered the establishment of an in-house repository, PharmTargetDB (unpublished results), which hosts pharmacophore models extracted from potential targets (co-complexed with corresponding small compounds) with available 3D structures. One of the purposes of this pharmacophore database initiative is to provide a pool of potential targets information for ‘target fishing’ with pharmacophore mapping method.
Herein, we present the first web-based tool PharmMapper for potential drug target prediction against any given small molecules via a ‘reverse’ pharmacophore mapping approach. The small molecule might be a biologically active compound detected in a cell- or animal-based bioassay screen, a natural product or an existing drug whose molecular target(s) is (are) unidentified. Benefited from the highly efficient and robust mapping method, PharmMapper bears high-throughput ability and can identify the potential target candidates from the database with a runtime of a few hours. Backed up by a large, in-house repertoire of pharmacophore database (PharmTargetDB) annotated with target information, PharmMapper may serve as a valuable tool for identifying targets for a novel synthetic compound, a newly isolated natural product, a compound with known biological activity or an existing drug whose mechanism of action is unknown.
METHODS
Construction of potential targets pharmacophore databases
PharmMapper requires a sufficient number of available pharmacophore models describing the binding modes of known ligands at the binding sites of protein targets. The target protein structures co-complexed with small molecules were carefully selected from DrugBank (21), BindingDB (22), PDBBind (23) and our PDTD (18) databases. DrugBank hosts a complete list of known targets with appropriate annotations, while BindingDB and PDBBind provide public, web-accessible databases of measured binding affinities, focusing chiefly on the interactions of those proteins considered to be drug targets with small or drug-like molecules. Only those proteins with available 3D crystal structures were selected and used for pharmacophore model extraction.
LigandScout, which is a software tool that allows rapid extraction of 3D pharmacophores from structural data of macromolecule–ligand complexes in a fully automated and convenient way (19), was used in the process of pharmacophore model derivation. Six primary types of pharmacophore features were adopted in this process: hydrophobic center (H), positive-charged center (P), negative-charged center (N), hydrogen bond acceptor vector (HBA), hydrogen bond donor vector (HBD) and aromatic plane (AR) and one optional feature [metal interaction center (M)]. Each ligand binding site was manually analyzed after generation of corresponding pharmacophore model and the corresponding shape was characterized by several excluded volumes centered at each residue of the binding pocket. All the small ligands with molecular weight lower than 100, such as solvents, buffers and metal cations, and all the cofactors with molecular weight over 600, such as CoAs, polypeptides and nucleic acids were regarded as ‘environment atoms’ instead of binding ligands. In this context, the corresponding pharmacophore models were not generated. For the proteins existing as homopolymers, only one monomer was reserved for analysis. For the proteins determined by NMR with multiple structure models, only the first model was selected for pharmacophore generation. As a result, we generated 7302 pharmacophore models (2241 entries are annotated as ‘Human protein targets’) and deposited them in PharmTargetDB. The target annotations were extracted from DrugBank, PDBSum (24), UniProt (25) and in-house TargetBank (our unpublished data) and were categorized as follows: UniProt access ID, target name, target function and indication/disease involved.
Reverse pharmacophore mapping procedure using PharmMapper
PharmMapper consists of two parts: a front-end web interface written in both PHP and HTML, with MySQL as database system, and a back-end tool for reverse pharmacophore mapping. The reverse pharmacophore mapping procedure is as follows: (i) PharmMapper flexibly aligns the given small molecule onto each pharmacophore model of proteins in the target list, and the fit values between the small molecule and the pharmacophores are calculated and recorded; (ii) PharmMapper presents the aligned pose with the corresponding pharmacophore model and prioritizes candidate targets based on the fit values to analyze the reverse mapping result. In general, PharmMapper outputs the top N hits of the ranking list, from which the user may select protein candidates for further bioassay validation.
Generally, the algorithm suggests to solve the molecule pharmacophore best fitting task in a strategy of sequential combination of triangle hashing (TriHash) and genetic algorithm (GA) optimization, which consists of following major steps: (i) ligand initialization and preparation; (ii) ligand as well as target pharmacophore model features triangulation; (iii) pairwise alignment and GA post optimization; and (iv) solution filtering, ranking and output. The readers can refer to the Supplementary Data for more details about the pharmacophore mapping algorithm used by PharmMapper.
WEB SERVER
PharmMapper server is open-accessed and free of charge. Users are expected to upload the mol2 file of the test molecule, customize the mapping parameters and submit a job. A job identity number, namely the JOB ID, is assigned to each job by the web server, and the number is appended to a job queue in the back-end server. The user may use the JOB ID to check the status of the submitted job.
Input
PharmMapper’s interface is very simple. Its input form has only one mandatory field: a file with single drug-like molecule or natural product stored in Mol2 format. The user must make sure the uploaded molecule has appropriate 3D structural information. Multiple commercial or open source toolkits are recommended to complete this task, including CORINA (26), CONCORD (27) and ChemAxon’s Standardizer (www.chemaxon.com). The user can choose or not to leave an email address in order to receive a notification when the job is finished. After uploading the file, the user is encouraged to set some optional parameters in the following pop-up form instead of accepting corresponding default values to reduce the computational cost or achieve more accurate result. Since PharmMapper uses semi-flexible alignment strategy, a conformer ensemble has to be generated prior to mapping. For single 3D conformer provided by the user, an in-house program Cyndi is used by default to generate multiple conformations. Of course, the user can skip this step by uploading pre-generated conformation ensemble with other programs, such as CAESAR (www.accelrys.com), MacroModel (www.schrodinger.com) and Omega (www.eyesopen.com). Additionally, the user can specify the minimum number of each pharmacophore feature type to skip those target pharmacophore models, of which the number of corresponding pharmacophore features are less than the threshold values. Moreover, the scoring weights assigned to each type of pharmacophore feature can be adjusted according to the user’s judgment towards the structural, physicochemical features presented by the molecule (e.g. if the molecule bears dominantly hydrophobic features, the scoring weight assigned to the Hydrophobic Score can be moderately increased to favor the hydrophobic interaction with the pharmacophore models). Detailed explanations for each field can be displayed in the pop-up windows when the mouse is lifted on the corresponding field and are also available in the Help page.
Output
A typical run of PharmMapper task takes 1–2 h, depending on the flexibility of the input molecule and filter parameters assigned by the user. To ensure successful job submission, the user is prompted to activate a self-refreshed alert page to monitor the job status. The user can bookmark this alert page so as to check the status of corresponding job at any time in the feature. Once the job completes, the user is automatically redirected to the computational results via the self-refreshed page or expected to input the JOB ID in the ‘Get Result’ page to access the computational results. The hyperlink to the result page is also contained in the notification after the job is finished, if the users have left their email address during job submission. The result will be kept on the server for up to 3 months so that the user may access the result at any time later via the same JOB ID.
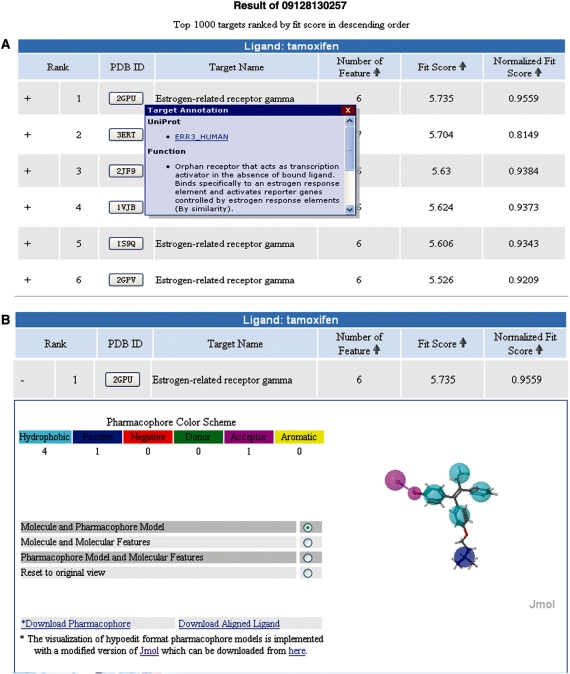
The output of a PharmMapper run is demonstrated in the form of a ranked list of hit target pharmacophore models that are sorted by fit score in descending order (Figure 1A). User can also re-rank the result list by normalized fit score or number of pharmacophore features in descending order via clicking the arrow icons in the corresponding columns. The 3D structural information can be accessed via the hyperlinks in the ‘PDB ID’ column to the Protein Database Bank (PDB) website (28). The hotlink to UniProt database as well as functional and therapeutic annotations of each target will be presented in the pop-up window by mouse lifting over the corresponding PDB IDs. As Figure 1B shows, a pull-down window will appear by clicking the ‘+’ mark at the starting of each line of the result table, which illustrates the details of each pharmacophore model candidate, including the numbers of each pharmacophore feature (rendered in different colors scheme), a 3D interactive visualization of molecule-pharmacophore alignment poses displayed via a modified version of Jmol applet (http://www.jmol.org), and the download links of the aligned pose of molecule as well as the corresponding pharmacophore model (in hypoedit format). The radio buttons in the pull-down window allow the users to show/hide either the pharmacophore model, query molecular conformation or the features from the query molecule in display, which may provide better visual assessment for the matching quality between the input probe molecule and the identified potential target pharmacophore models. All the text-based targets information is downloadable in comma separated values (CSV) format via the hotlink at the bottom of the result page.
Figure 1.
An example of the output of PharmMapper. (A) The ranked list of hit target pharmacophore models, which are sorted by fit score in descending order. (B) The pull-down window that illustrates the details of each pharmacophore model candidate and the molecule pharmacophore alignment.
TEST CASES
To test the reliability of the PharmMapper server, the potential drug target proteins for tamoxifen were searched via PharmMapper server. The result and its comparison with the published experimental data are described below. Another test case to identify the potential targets of methotrexate is presented in the Supplementary Table S3.
Tamoxifen is used as an adjuvant therapy in the treatment of breast cancer (29). It has been proved as a multiple target drug. So far, 14 proteins have been identified as interaction targets for tamoxifen or 4H-tamoxifen, which is the active metabolite of tamoxifen (30–41; Supplementary Data Table S1). The top 1000 (actually 912 hits) pharmacophore candidates identified via PharmMapper are listed in Supplementary Table S2 and those corresponding to the proteins identified by experimental data are shown in Table 1. Four among the top 100 candidates are annotated as known targets of tamoxifen, namely estrogen receptor (Rank 1), 17β-hydroxysteroid dehydrogenase (Rank 18), dihydrofolate reductase (Rank 29) and glutathione transferase (Rank 49). The top 300 candidates include six additional targets identified experimentally, i.e. prostaglandin synthase (124), collagenase (Rank 136), carboxylesterase 1 (Rank 130), 3α-hydroxysteroid dehydrogenase (Rank 168), protein kinase C (Rank 222) and calmodulin (Rank 297). Another tamoxifen target (alcohol dehydrogenase) is ranked 817. Of experimentally confirmed targets for tamoxifen, 29% and 71% appear among the top 100 and 300 of the PharmMapper predicted candidates, respectively, and 11 of the 14 experimentally confirmed tamoxifen targets are covered in the top 1000 pharmacophore models, indicating the reliability of this server tool.
Table 1.
Pharmacophore candidates of tamoxifen identified by PharmMapper
| Rank | PDB ID | Fit score | Target name |
|---|---|---|---|
| 1 | 2EWP | 5.770 | Estrogen-related receptor-γ |
| 18 | 1I5R | 4.121 | Estradiol 17-β-dehydrogenase 1 |
| 29 | 1DG7 | 3.796 | Dihydrofolate reductase |
| 49 | 1GSF | 3.755 | Glutathione S-transferase A1 |
| 124 | 1PXX | 3.407 | Prostaglandin G/H synthase 2 |
| 130 | 1YA4 | 3.395 | Liver carboxylesterase 1 |
| 136 | 3I7I | 3.383 | Collagenase 3 |
| 168 | 1HDC | 3.309 | 3-α (or 20-β)-hydroxysteroid dehydrogenase |
| 222 | 1XJD | 3.118 | Protein kinase C-θ type |
| 297 | 1XA5 | 2.950 | Calmodulin |
| 817 | 1MGO | 2.885 | Alcohol dehydrogenase E chain |
A receiver operating characteristic (ROC) depicts the fraction of true positives versus the fraction of false positives found in a classification experiment. The area under the ROC curve (AUC) equals the probability of ranking a randomly selected true target higher than a randomly selected decoy target. Ideal distributions of true targets and decoys result in an AUC value approaching 1.0, whereas random distributions result in a value of 0.5. The ROC enrichment (ROCE) expresses the percentage of true targets observed as a proportion of the percentage of the decoy targets observed, and ROCE values of >1.0 signify enrichment with respect to random distributions. The AUC and ROCE at four decoy levels were used as the performance metric of PharmMapper in the benchmark test for tamoxifen target identification. Since there is no other available pharmacophore-based drug identification method using the same drug target database as we used, only PharmMapper’s result is presented in Table 2. The AUC value is 0.7 and the ROCE value at 0.5% decoy achieves 28.7, which is promising and reliable for a retrospective target identification case.
Table 2.
AUC value and ROCE values at 0.5, 1 and 2% decoy levels for the case of tamoxifen
| True targets/decoy | AUC | ROCE |
|||
|---|---|---|---|---|---|
| 0.5% | 1% | 2% | 5% | ||
| 216/7069 | 0.70 | 28.7 | 14.82 | 8.80 | 4.54 |
SUMMARY
We presented here the first web server for potential drug targets identification via large-scale reverse pharmacophore mapping strategy. The abundant potential target entries represented by pharmacophore models in the repository of PharmTargetDB, and moreover, the efficient pharmacophore mapping algorithm behind the server allow the fast and reliable identification of the pharmacophore target candidates for small molecules such as drugs, lead compounds and natural products. The user interface is very simple and the algorithm is fully automated, because the user is only required to upload one Mol2 file containing the 3D information of the query molecule. In addition, for experienced users, more optional parameters for speedy and accurate control as well as the candidate target subset to be searched can be freely customized. The intuitive and interactive mode for results display allows in situ manual validation of the predicted aligned poses between the query molecule and corresponding pharmacophore model hits. The validation example of multiple potential targets identification for tamoxifen illustrated that PharmMapper can provide useful clues for further bioassay in drug–target interaction research.
As a similar and complementary server of TarfisDock, PharmMapper can also be used in mapping the regulation genomic network for an existing drug or a drug candidate, as well as in profiling the potential secondary or side effects for a drug molecule in a different viewpoint from the regular chemogenetic method. These are useful clues for further experimental test in evaluating the efficacy and toxicity of the drug. On the other hand, the target information produced by PharmMapper is also significant for functional genomic study within the chemical biology paradigm. Currently, a web-based screening platform to find lead compounds with PharmMapper, allowing customized and selected pharmacophore from the PharmTargetDB, is underway.
To some extent, PharmMapper still has some limitations: the pharmacophore database only includes drug targets that have PDB structures with co-crystallized ligand. Thus, some potential targets of the query ligand could be missed due to the limited coverage of the database. However, as the number of structures deposited in PDB grows exponentially yearly, the PharmTargetDB we used is updated periodically and new targets whose complex structures are released can be added to extend the database coverage. We are also collecting the ligand-based pharmacophore models built in-house or reported by other group for the important drug targets without crystal structures like G protein-coupled receptors and ion channels to cover more targets information in PharmTargetDB.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Major State Basic Research Project (grants 2009CB918501 and 2009CB918502); National Natural Science Foundation of China (grants 20803022 and 20721003); Shanghai Committee of Science and Technology (grants 09dZ1975700 and 08JC1407800); 863 Hi-Tech Program of China (grants 2007AA02Z304 and 2007AA02Z330); Major National Scientific and Technological Project of China (grants 2009ZX09501-001 and 2009ZX09301-001); 111 Project (grant B07023); Shanghai Rising-Star Program (grant 10QA1401800 to H.L.). Funding for open access charge: the 863 Hi-Tech Program of China (grant 2007AA02Z304).
Conflict of interest statement. None declared.
Supplementary Material
REFERENCES
- 1.Hopkins AL, Groom CR. The druggable genome. Nat. Rev. Drug Discov. 2002;1:727–730. doi: 10.1038/nrd892. [DOI] [PubMed] [Google Scholar]
- 2.Overington JP, Al-Lazikani B, Hopkins AL. How many drug targets are there? Nat. Rev. Drug Discov. 2006;5:993–996. doi: 10.1038/nrd2199. [DOI] [PubMed] [Google Scholar]
- 3.Bajorath J. Computational analysis of ligand relationships within target families. Curr. Opin. Chem. Biol. 2008;12:352–358. doi: 10.1016/j.cbpa.2008.01.044. [DOI] [PubMed] [Google Scholar]
- 4.Huang CM, Elmets CA, Tang DC, Li F, Yusuf N. Proteomics reveals that proteins expressed during the early stage of Bacillus anthracis infection are potential targets for the development of vaccines and drugs. Genomics, Proteomics Bioinformatics. 2004;2:143–151. doi: 10.1016/S1672-0229(04)02020-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Klebe G. Virtual ligand screening: strategies, perspectives and limitations. Drug Discov. Today. 2006;11:580–594. doi: 10.1016/j.drudis.2006.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.O’Connor KA, Roth BL. Finding new tricks for old drugs: an efficient route for public-sector drug discovery. Nat. Rev. Drug Discov. 2005;4:1005–1014. doi: 10.1038/nrd1900. [DOI] [PubMed] [Google Scholar]
- 7.Keiser MJ, Roth BL, Armbruster BN, Ernsberger P, Irwin JJ, Shoichet BK. Relating protein pharmacology by ligand chemistry. Nat. Biotechnol. 2007;25:197–206. doi: 10.1038/nbt1284. [DOI] [PubMed] [Google Scholar]
- 8.Campillos M, Kuhn M, Gavin AC, Jensen LJ, Bork P. Drug target identification using side-effect similarity. Science. 2008;321:263–266. doi: 10.1126/science.1158140. [DOI] [PubMed] [Google Scholar]
- 9.Bender A, Young DW, Jenkins JL, Serrano M, Mikhailov D, Clemons PA, Davies JW. Chemogenomic data analysis: prediction of small-molecule targets and the advent of biological fingerprint. Comb. Chem. High Throughput Screen. 2007;10:719–731. doi: 10.2174/138620707782507313. [DOI] [PubMed] [Google Scholar]
- 10.Jacoby E. Chemogenomics: drug discovery’s panacea? Mol. Biosyst. 2006;2:218–220. doi: 10.1039/b603004c. [DOI] [PubMed] [Google Scholar]
- 11.Mestres J. Computational chemogenomics approaches to systematic knowledge-based drug discovery. Curr. Opin. Drug Discov. Devel. 2004;7:304–313. [PubMed] [Google Scholar]
- 12.Rognan D. Chemogenomic approaches to rational drug design. Br. J. Pharmacol. 2007;152:38–52. doi: 10.1038/sj.bjp.0707307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Savchuk NP, Balakin KV, Tkachenko SE. Exploring the chemogenomic knowledge space with annotated chemical libraries. Curr. Opin. Chem. Biol. 2004;8:414–417. doi: 10.1016/j.cbpa.2004.06.003. [DOI] [PubMed] [Google Scholar]
- 14.Wuster A, Madan BM. Chemogenomics and biotechnology. Trends Biotechnol. 2008;26:252–258. doi: 10.1016/j.tibtech.2008.01.004. [DOI] [PubMed] [Google Scholar]
- 15.Hert J, Keiser MJ, Irwin JJ, Oprea TI, Shoichet BK. Quantifying the relationships among drug classes. J. Chem. Inf. Model. 2008;48:755–765. doi: 10.1021/ci8000259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xie L, Li J, Xie L, Bourne PE. Drug discovery using chemical systems biology: identification of the protein-ligand binding network to explain the side effects of CETP inhibitors. PLoS Comput. Biol. 2009;5:e1000387. doi: 10.1371/journal.pcbi.1000387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li HL, Gao ZT, Kang L, Zhang HL, Yang K, Yu KQ, Luo XM, Zhu WL, Chen KX, Shen JH, et al. TarFisDock: a web server for identifying drug targets with docking approach. Nucleic Acids Res. 2006;34:W219–224. doi: 10.1093/nar/gkl114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gao ZT, Li HL, Zhang HL, Liu XF, Kang L, Luo XM, Zhu WL, Chen KX, Wang XC, Jiang HL. PDTD: a web-accessible protein database for drug target identification. BMC Bioinformatics. 2008;9:104. doi: 10.1186/1471-2105-9-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wolber G, Langer T. LigandScout: 3-D pharmacophores derived from protein-bound ligands and their use as virtual screening filters. J. Chem. Inf. Model. 2005;45:160–169. doi: 10.1021/ci049885e. [DOI] [PubMed] [Google Scholar]
- 20.Steindl M, Schuster D, Wolber G, Laggner C, Langer C. High throughput structure-based pharmacophore modeling as a basis for successful parallel virtual screening. J. Comput. Aided Mol. Des. 20:703–715. doi: 10.1007/s10822-006-9066-y. [DOI] [PubMed] [Google Scholar]
- 21.Wishart DS, Knox C, Guo AC, Cheng D, Shrivastava S, Tzur D, Gautam B, Hassanali M. DrugBank: a knowledgebase for drugs, drug actions and drug targets. Nucleic Acids Res. 2008;36:D901–906. doi: 10.1093/nar/gkm958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu T, Lin Y, Wen X, Jorissen RN, Gilson MK. BindingDB: a web-accessible database of experimentally determined protein-ligand binding affinities. Nucleic Acids Res. 2007;35:D198–201. doi: 10.1093/nar/gkl999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang R, Fang X, Lu Y, Wang S. The PDBbind database: collection of binding affinities for protein-ligand complexes with known three-dimensional structures. J. Med. Chem. 2004;47:2977–2980. doi: 10.1021/jm030580l. [DOI] [PubMed] [Google Scholar]
- 24.Laskowski RA, Hutchinson EG, Michie AD, Wallace AC, Jones ML, Thornton JM. PDBsum: a Web-based database of summaries and analyses of all PDB structures. Trends Biochem. Sci. 1997;22:488–490. doi: 10.1016/s0968-0004(97)01140-7. [DOI] [PubMed] [Google Scholar]
- 25.UniProt Consortium. The universal protein resource (UniProt) Nucleic Acids Res. 2007;35:D193–197. doi: 10.1093/nar/gkl929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sadowski J, Gasteiger J. From atoms and bonds to 3-dimensional atomic coordinates - automatic model builders. Chem. Rev. 1993;93:2567–2581. [Google Scholar]
- 27.Sheridan RP, Rusinko A, Nilakantan R, Venkataraghavan R. Searching for pharmacophores in large coordinate data bases and its use in drug design. Proc. Natl Acad. Sci. USA. 1989;86:8165–8169. doi: 10.1073/pnas.86.20.8165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The protein data bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hughes-Davies L, Caldas C, Wishart GC. Tamoxifen: the drug that came in from the cold. Br. J. Cancer. 2009;101:875–878. doi: 10.1038/sj.bjc.6605231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Favoni RE, de Cupis A. Steroidal and nonsteroidal oestrogen antagonists in breast cancer: basic and clinical appraisal. Trends Pharmacol. Sci. 1998;19:406–415. doi: 10.1016/s0165-6147(98)01252-8. [DOI] [PubMed] [Google Scholar]
- 31.Rowlands MG, Budworth J, Jarman M, Hardcastle IR, McCague R, Gescher A. Comparison between inhibition of protein kinase C and antagonism of calmodulin by tamoxifen analogues. Biochem. Pharmacol. 1995;50:723–726. doi: 10.1016/0006-2952(95)00186-4. [DOI] [PubMed] [Google Scholar]
- 32.Nilsson UW, Garvin S, Dabrosin C. MMP-2 and MMP-9 activity is regulated by estradiol and tamoxifen in cultured human breast cancer cells. Breast Cancer Res. Treat. 2007;102:253–261. doi: 10.1007/s10549-006-9335-4. [DOI] [PubMed] [Google Scholar]
- 33.Messiha FS. Leu-enkephalin, tamoxifen and ethanol interactions: effects on motility and hepatic ethanol metabolizing enzymes. Gen. Pharmacol. 1990;21:45–48. doi: 10.1016/0306-3623(90)90593-b. [DOI] [PubMed] [Google Scholar]
- 34.Santner SJ, Santen RJ. Inhibition of estrone sulfatase and 17beta-hydroxysteroid dehydrogenase by antiestrogens. J. Steroid Biochem. Mol. Biol. 1993;45:383–390. doi: 10.1016/0960-0760(93)90007-j. [DOI] [PubMed] [Google Scholar]
- 35.Ritchie GA. The direct inhibition of prostaglandin synthetase of human breast cancer tumor tissue by tamoxifen. Recent Results Cancer Res. 1980;71:96–101. doi: 10.1007/978-3-642-81406-8_13. [DOI] [PubMed] [Google Scholar]
- 36.Lax ER, Rumstadt F, Plasczyk H, Peetz A, Schriefers H. Antagonistic action of estrogens, flutamide, and human growth hormone on androgen-induced changes in the activities of some enzymes of hepatic steroid metabolism in the rat. Endocrinology. 1983;113:1043–1055. doi: 10.1210/endo-113-3-1043. [DOI] [PubMed] [Google Scholar]
- 37.Nuwaysir EF, Daggett DA, Jordan VC, Pitot HC. Phase II enzyme expression in rat liver in response to the antiestrogen tamoxifen. Cancer Res. 1996;56:3704–3710. [PubMed] [Google Scholar]
- 38.Levine RM, Rubalcaba E, Lippman ME, Cowan KH. Effects of estrogen and tamoxifen on the regulation of dihydrofolate reductase gene expression in a human breast cancer cell line. Cancer Res. 1985;45:1644–1650. [PubMed] [Google Scholar]
- 39.Paavonen T, Aronen H, Pyrhonen S, Hajba A, Andersson LC. The effect of toremifene therapy on serum immunoglobulin levels in breast cancer. Apmis. 1991;99:849–853. doi: 10.1111/j.1699-0463.1991.tb01270.x. [DOI] [PubMed] [Google Scholar]
- 40.de-Médina P, Favre G, Poirot M. Multiple targeting by the antitumor drug tamoxifen: a structure-activity study. Curr. Med. Chem. Anticancer Agents. 2004;4:491–508. doi: 10.2174/1568011043352696. [DOI] [PubMed] [Google Scholar]
- 41.Fleming CD, Bencharit S, Edwards CC, Hyatt JL, Tsurkan L, Bai F, Fraga C, Morton CL, Howard-Williams EL, Potter PM, et al. Structural insights into drug processing by human carboxylesterase 1: tamoxifen, mevastatin, and inhibition by benzil. J. Mol. Biol. 352:165–177. doi: 10.1016/j.jmb.2005.07.016. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.