Abstract
The WHAT IF molecular-modelling and drug design program is widely distributed in the world of protein structure bioinformatics. Although originally designed as an interactive application, its highly modular design and inbuilt control language have recently enabled its deployment as a collection of programmatically accessible web services. We report here a collection of WHAT IF-based protein structure bioinformatics web services: these relate to structure quality, the use of symmetry in crystal structures, structure correction and optimization, adding hydrogens and optimizing hydrogen bonds and a series of geometric calculations. The freely accessible web services are based on the industry standard WS-I profile and the EMBRACE technical guidelines, and are available via both REST and SOAP paradigms. The web services run on a dedicated computational cluster; their function and availability is monitored daily.
INTRODUCTION
Macromolecular structures are at the basis of much research in fields as diverse as drug design, bio-fuel engineering, molecular biology or force field design. The number of new protein structures deposited in the PDB has remained stable for several years at around 20 per day. The statistics on web servers listed in the special volumes of NAR collected by Brazas et al. (1) clearly show that protein structures remain an important topic, as the number of web servers and visualization tools relating to protein structures continues to grow steadily. WHAT IF (2) is a widely distributed interactive software package for macromolecular analysis, visualization, modelling and structure validation. Originally an interactive application driven either via a Graphical User Interface or from the command line, a decade ago we exposed the first three components of WHAT IF via human-accessible web interfaces (3). Since then, the collection of functions accessible in this way has grown to around 80, which are being used by a few hundred users each day. More recently, the software has been successfully integrated with the COOT crystallographic software (4) and the YASARA (http://www.yasara.org/) molecular modelling, simulation and drug design software (5).
Encouraged by initiatives such as BioSapiens (http://www.biosapiens.info/) and EMBRACE (http://www.embracegrid.info/), which have championed the use of web services in the life sciences, we have now developed and deployed a freely available web service version of WHAT IF. Unlike the more traditional web server (e.g. HTML/CGI) interfaces that have become a popular way of providing remote human access to tools and resources, web services are straightforwardly accessible from within computer programs, allowing them to be used for large-scale ‘batch processing’ tasks that would be inconceivable via manual cut-and-paste alternatives. Acting as a subroutine or function that resides on a remote computer, web services provide a number of benefits to bioinformatics software developers (and thence directly or indirectly also to life scientist users of their tools):
reduction in the amount of code that is being re-invented and maintained;
reduction in the cost of installing, managing and maintaining local installations of software packages;
access to the latest version of algorithms and data without the need to upgrade local installations; and
access to enormous amounts of (free) CPU time.
A more extensive description of the advantages and disadvantages of the use of web services can be found in the article describing the EMBRACE web service registry elsewhere in this volume. The first WHAT IF-based web servers were launched in 1997 and were published in 1998 (3) and remain active to this day (http://swift.cmbi.ru.nl/).
METHODS
WHAT IF, a FORTRAN program conceived in 1987 (2), is still actively maintained and developed. Its core of around a million lines of code represent a substantial collection of algorithms and heuristics that remain of significant utility to today’s scientific community. In order to deploy the WHAT IF code as a web service, we designed a special ‘wrapper’ application that acts as an intermediate between the legacy architecture of the command-line-based WHAT IF system and the modern protocols required by web services. This application, called WIWS (‘WHAT IF web services’), listens on the network for incoming web service requests and translates these into WHAT IF commands that are injected into the interactive component of the system as though they had been typed by a user. WHAT IF then executes these commands and produces its own bespoke format output, which in turn is then parsed by WIWS, and translated back to the contemporary protocols (e.g. SOAP and XML) expected by web service software. The WSDL (‘Web Service Description Language’—a formal description of the web service, required by client software in order to access the service) and the documentation for the web services are generated automatically by WIWS and are available at http://wiws.cmbi.ru.nl/wsdl/ and http://wiws.cmbi.ru.nl/help/, respectively. Experienced WHAT IF users can freely obtain from us all software elements needed to set up their own WHAT IF-based web services.
WHAT IF has over 1600 different functions that can be invoked using a command line interface, and which in essence form a simple state-based programming language. Rather than expose all of this complex fine-grained level of control as web services, we have instead developed a scripting language that allows more meaningful course-grained web services to be automatically generated by combining WHAT IF’s low-level commands in a coherent manner.
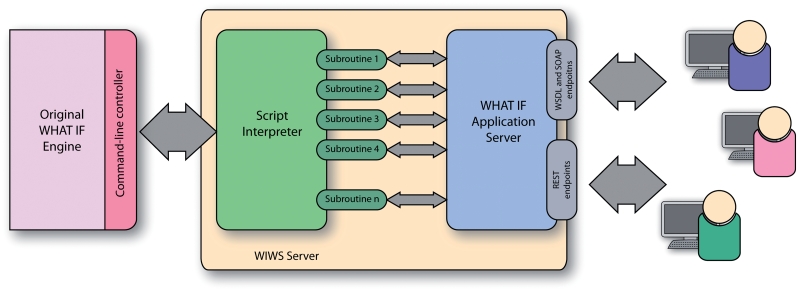
The architecture of the system (Figure 1) thus consists of two components: (i) the original WHAT IF core, acting as an engine responsible for executing the underlying algorithms; (ii) the WIWS system: this contains (a) an interpreter and scripts containing subroutines that map between sequences of low-level WHAT IF commands and meaningful web service-level operations, and (b) a bespoke HTTP server that responds to incoming web service requests and invokes the high-level functions represented by individual subroutines.
Figure 1.
Architecture of the WIWS system. The original WHAT IF system at the far left is augmented with a software layer that receives streams of automated commands, and sends them to WHAT IF as though they had been typed at the command line. The WIWS server (centre) receives Web service invocations from users in either REST or SOAP format, and matches these with one of the scripted subroutines, which then calls WHAT IF in order to execute the appropriate sequence of instructions.
When the server is initialized, it parses the latest version of its script and automatically generates and advertises the WSDL descriptions of the subroutines described therein, as well as creating corresponding SOAP and REST endpoints. The server is written in C++. The script language is based on the Oberon-2 programming language and implements most of its syntax. The XML and HTTP handling code comes from the open source libzeep project (http://libzeep.berlios.de/).
RESULTS
To date, we have produced 84 web servers and 64 web services, all of which take PDB format molecular structure data as input. The web servers mainly deal with options that produce two- and three-dimensional graphics and other WHAT IF options that produce human readable output. The web services focus on WHAT IF options that would be useful for third-party protein structure-related software tools. The web service operations broadly fall into one of two groups. In the first group are operations that categorize or otherwise annotate residues or atoms, for example to:
provide simple geometric parameters, such as accessibilities, torsion angles, etc.;
list observables, such as disulphide bridges, salt bridges, etc.;
provide tables of per-residue structure validation scores.
In the second group are operations that generate, or otherwise modify, the input’s molecular structure, for example to:
correct and or complete coordinates;
add and optimize protons; and
add symmetry-related residues/molecules.
A representative selection of these services is shown in Table 1.
Table 1.
Selected WHAT IF web services (of the 64 that are available to date)
| Visualization options | What the web service returns: |
|---|---|
| GetSurfaceDot | (Many) points in space that visualize the surface. |
| Geometric calculations | |
| AtomAccessibilitySolvent | Solvent-accessible surface area for each atom in Ångström2. |
| ResidueAccessibilityVacuum | Solvent-accessible surface area in Ångström2, for the residue in vacuum, only including the backbone of its direct neighbours. |
| ResidueTorsions | For each residue, its torsion angles: φ, ψ, ω, χ1–χ5 (in degrees). |
| ShowTauAngle | For each amino acid, its backbone τ-angle (in degrees). |
| CysteineTorsions | Cys-cys bridge torsions over: Cαi-Cβi Cβi-Sγi Sγi-Sγj Sγj-Cβj Cβj-Cαj |
| Atomic contacts | |
| ShowBumps | Atom pairs with a clash worse than 0.25 Å. |
| ShowSaltBridges | Pairs of charged groups within 8.0 Å. |
| ShowHydrogenBonds | H-bonded atoms: four geometric parameters describe the bond. |
| ShowLigandContacts | Lists macromolecule-ligand contacting atom pairs. |
| ShowCysteineMetal | Lists cysteines that are bound to a metal. |
| HasNucleicContacts | Lists protein residues that contact a nucleic acid. |
| Structure quality | |
| PackingQuality | Packing normality for amino acids. |
| ImproperQualityMax | Per residue, its worst improper dihedral Z-score. |
| Protein engineering | |
| ShowLikelyRotamers | Show rotamer likelihoods for rotamers for all amino acid types. |
| ProlineMutationValue | Likelihood at each position that an introduced proline will be thermostabilizing. |
| Returning coordinates | |
| PDBasXML | The input PDB file is returned in XML. |
| CorrectedPDBasXML | The input PDB gets 22 correction steps and is returned as XML. |
| SymShellFiveXML | Shell of symmetry-related residues, 5.0 Å thick, as XML. |
The full list and all descriptions are available at http://wiws.cmbi.ru.nl/help/. The algorithms underlying the web services are explained at the WHAT IF web site (http://swift.cmbi.ru.nl/whatif/).
All the WHAT IF web services are regularly tested by a script that exercises them against 10 valid PDB files that have been selected for their complexity (from a WHAT IF point of view). The services have also been entered into the EMBRACE registry (6), which regularly monitors and reports on their availability. Usage of the web services is free and essentially unlimited, but we request that users submit their web service calls sequentially.
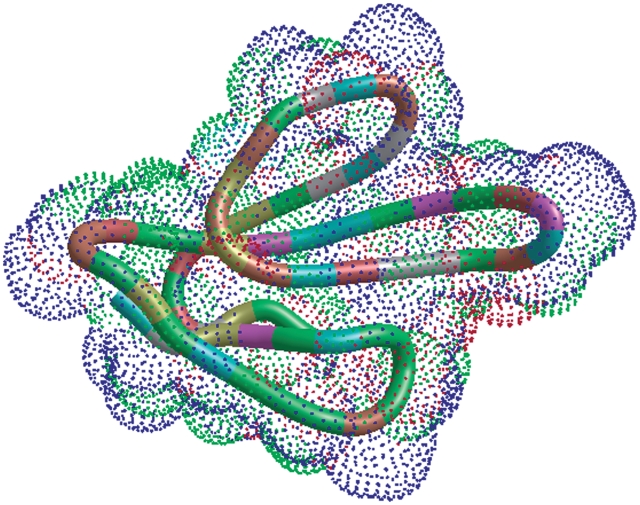
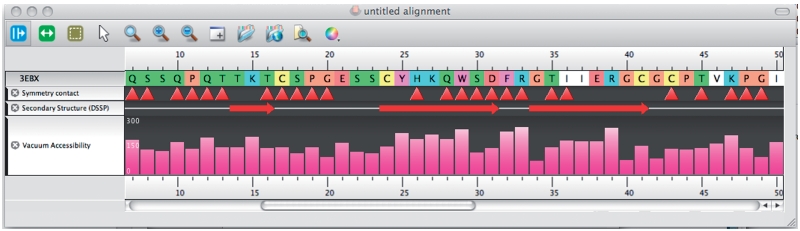
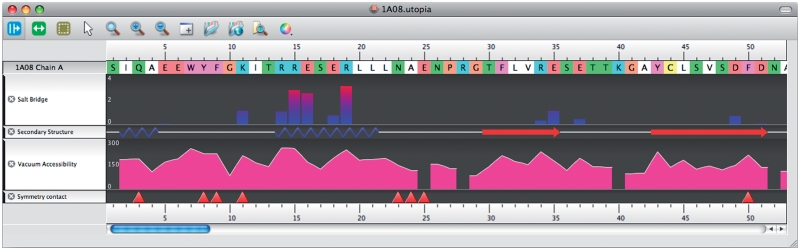
As a case study, 10 of the WHAT IF web service operations have been integrated with the Utopia (7,8) visualization suite. Utopia has a flexible plug-in system that allows its graphical visualization components to be linked easily with online tools and databases. Relevant fragments of the Python code necessary to use the WHAT IF web services in Utopia are listed at the WHAT IF (http://swift.cmbi.ru.nl/whatif/) web service documentation page (http://swift.cmbi.ru.nl/Webservices/). Utopia is freely available from http://www.getutopia.com/. Figures 2 and 3 show Utopia screenshots that illustrate the utility of the web service-based virtual linkup of the two software packages. The integrated perspectives shown in the Figures provide at-a-glance overviews of biophysical characteristics both of the molecular structure and of individual amino acids, and how these interrelate: this facilitates, for example, analysis of crystal-packing contact residues relative to core secondary structures (e.g. contact residues 26–36 in Figure 2 lie on the long projecting finger-like loop in the centre of the 3D structure); identification of the locations and relative strength of salt-bridging residues, again in the context of the secondary structures in which they are located; and so on (Figures 3 and 4).
Figure 2.
Left, a representation PDB file 3EBX (9,10). The all-atom dot surface is calculated by WHAT IF, and is colour coded by atom type. The backbone ribbon, generated by Utopia, is coloured according to residue physicochemical characteristics: green, polar neutral; red, polar acidic; blue, polar basic; white, hydrophobic aliphatic; purple, hydrophobic aromatic; brown, special structural (proline, glycine); yellow, sulphur-containing, structural (cysteine).
Figure 3.
Here, the horizontal tracks show in order (i) the amino acid residue number in the sequence, (ii) the sequence itself (colour code as above), (iii) residues with crystal-packing contacts (indicated by red triangles); (iv) the secondary structure of 3EBX determined by DSSP (11), filtered by WHAT IF and displayed by Utopia—here, red arrows denote β-strands; (v) vacuum accessibility in square Ångströms.
Figure 4.
PDB file 1A08 (12) displayed using Utopia, having invoked 4 WHAT IF services. Tracks show in order: (i) amino acid residue number, (ii) the residue sequence (colour code as in Figure 3); (iii) residues involved in salt bridges (height of the bar indicates the total enthalpy contribution of all salt bridges in which the residue is involved); (iv) secondary structure (blue zigzags indicate α-helices, red arrows denote β-strands); (v) vacuum accessibility displayed using an alternative mode of Utopia visualization relative to that in Figure 3 (the values at 25, 28, 40 and 52 are missing because their side chains are not complete in the PDB file); (vi) red triangles indicate residues involved in crystal-packing contacts.
The use of the WHAT IF web services is rather straightforward. The WSDL will suffice for most experienced programmers, so we will list here only one small example, mainly to illustrate the ease of use of the WHAT IF web services. The smallest Python program we could think of that actually does something is listed below. The first line in this four-line program points at the Python executable. The second line imports the suds Python module (which you can download from http://python-suds.sourceforge.net/). The third line tells the program where the WHAT IF web services WSDL is located and the fourth line tells these services to print the results of the ShowBumps web service for the PDB file with identifier 1crn.
#!/usr/bin/python
from suds import client
c = client.Client(‘http://wiws.cmbi.ru.nl/wsdl’)
print c.service.ShowBumps(‘1crn')
DISCUSSION
We have produced 64 web services that perform a wide variety of macromolecular structure-related tasks. While selecting and designing these web services, we especially kept in mind the many programmers who provide macromolecular software and/or web services for niche research areas and who thus could benefit most from easy access to some of WHAT IF’s more advanced features, such as protein structure validation, structure correction and hydrogen bond- and symmetry application-related options. The automatic generation of web services via our scripting language means that deploying new services is a relatively trivial task, and we welcome and encourage requests from the community for new services.
FUNDING
NBIC, EMBRACE and UK BBSRC (grant number BBE0160651); EMBRACE project is funded by the European Commission within its FP6 Programme, under the thematic area ‘Life sciences, genomics and biotechnology for health’ (contract number LHSG-CT-2004-512092). Funding for open access charge: CMBI departmental money.
Conflict of interest statement. None declared.
REFERENCES
- 1.Brazas MD, Yamada JT, Ouellette BF. Evolution in bioinformatic resources: 2009 update on the Bioinformatics Links Directory. Nucleic Acids Res. 2009;37:W3–W5. doi: 10.1093/nar/gkp531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vriend G. WHAT IF: a molecular modeling and drug design program. J. Mol. Graph. 1990;8:52–56, 29. doi: 10.1016/0263-7855(90)80070-v. [DOI] [PubMed] [Google Scholar]
- 3.Rodriguez R, Chinea G, Lopez N, Pons T, Vriend G. Homology modeling, model and software evaluation: three related resources. Bioinformatics. 1998;14:523–528. doi: 10.1093/bioinformatics/14.6.523. [DOI] [PubMed] [Google Scholar]
- 4.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 5.Krieger E, Koraimann G, Vriend G. Increasing the precision of comparative models with YASARA NOVA–a self-parameterizing force field. Proteins. 2002;47:393–402. doi: 10.1002/prot.10104. [DOI] [PubMed] [Google Scholar]
- 6.Pettifer S, Thorne D, McDermott P, Attwood T, Baran J, Bryne JC, Hupponen T, Mowbray D, Vriend G. An active registry for bioinformatics web services. Bioinformatics. 2009;25:2090–2091. doi: 10.1093/bioinformatics/btp329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pettifer SR, Sinnott JR, Attwood TK. UTOPIA-User-Friendly Tools for Operating Informatics Applications. Comp. Funct. Genomics. 2004;5:56–60. doi: 10.1002/cfg.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pettifer S, Thorne D, McDermott P, Marsh J, Villeger A, Kell DB, Attwood TK. Visualising biological data: a semantic approach to tool and database integration. BMC Bioinformatics. 2009;10(Suppl. 6):S19. doi: 10.1186/1471-2105-10-S6-S19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Smith JL, Corfield PWR, Hendrickson WA, Low BW. Refinement at 1.4 A resolution of a model of erabutoxin b: treatment of ordered solvent and discrete disorder. Acta Crystallogr. A. 1988;44:357–368. doi: 10.1107/s0108767388000303. [DOI] [PubMed] [Google Scholar]
- 10.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kabsch W, Sander C. Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers. 1983;22:2577–2637. doi: 10.1002/bip.360221211. [DOI] [PubMed] [Google Scholar]
- 12.Charifson PS, Shewchuk LM, Rocque W, Hummel CW, Jordan SR, Mohr C, Pacofsky GJ, Peel MR, Rodriguez M, Sternbach DD, et al. Peptide ligands of pp60(c-src) SH2 domains: a thermodynamic and structural study. Biochemistry. 1997;36:6283–6293. doi: 10.1021/bi970019n. [DOI] [PubMed] [Google Scholar]