Abstract
DSAP is an automated multiple-task web service designed to provide a total solution to analyzing deep-sequencing small RNA datasets generated by next-generation sequencing technology. DSAP uses a tab-delimited file as an input format, which holds the unique sequence reads (tags) and their corresponding number of copies generated by the Solexa sequencing platform. The input data will go through four analysis steps in DSAP: (i) cleanup: removal of adaptors and poly-A/T/C/G/N nucleotides; (ii) clustering: grouping of cleaned sequence tags into unique sequence clusters; (iii) non-coding RNA (ncRNA) matching: sequence homology mapping against a transcribed sequence library from the ncRNA database Rfam (http://rfam.sanger.ac.uk/); and (iv) known miRNA matching: detection of known miRNAs in miRBase (http://www.mirbase.org/) based on sequence homology. The expression levels corresponding to matched ncRNAs and miRNAs are summarized in multi-color clickable bar charts linked to external databases. DSAP is also capable of displaying miRNA expression levels from different jobs using a log2-scaled color matrix. Furthermore, a cross-species comparative function is also provided to show the distribution of identified miRNAs in different species as deposited in miRBase. DSAP is available at http://dsap.cgu.edu.tw.
INTRODUCTION
Next-generation sequencing (NGS) technologies have found broad applicability in functional genomics research. The main advantage of NGS technologies is eliminating the need for in vivo cloning by clonal amplification of spatially separated single molecules using either emulsion PCR (Roche 454 and Applied Biosystems SOLiD) or bridge amplification on a solid surface (Illumina Solexa Genome Analyzer). NGS has been used extensively for expression profiling and discovery of microRNAs (miRNAs) and other small non-coding RNAs (ncRNAs) in many organisms. miRNAs are a growing family of regulatory molecules with several important biological functions involved in development, differentiation, proliferation, apoptosis and response to stress. Dysregulation of miRNA expression also contributes to disease pathology (1–4). Since the discovery of the first two miRNAs, lin-4 and let-7 (5,6), in the Nematode Caenorhabditis elegans, miRNAs have been described in invertebrates, vertebrates, plants, yeast and more recently in protists. The precursor miRNAs are cleaved in the nucleus by the Drosha enzyme to a 70 nucleotide hairpin transcript (pre-miRNA), transported to the cytoplasm by Exportin 5 through nuclear pores and then cleaved by Dicer (RNase III enzyme) into 19–22 nt double-stranded transcripts. In cytoplasm, the mature miRNA is loaded into an RNA-induced silencing complex (RISC) to form a miRNA-ribonucleoprotein complex (miRNP) and binds to the target sites of mRNAs (7–11) predominately in the untranslated region of the target mRNA for translational repression or mRNA cleavage (10–12).
Direct cloning and sequencing of small RNAs from organisms in the pre-NGS era was time consuming and expensive. The relatively low cost of NGS and the number of reads generated from a single run has brought the field of miRNA research back into the laboratories of single investigators, as is evidenced by the fact that the majority of publications on NGS of miRNA originated at sites other than the large genome centers. A major problem for large-scale massive parallel sequencing of miRNAs is the handling and analysis of generated data. NGS platforms can easily generate a gigabase of nucleotides per run, which is equivalent to the output of more than 50 Applied Biosystems 3730XL capillary sequencers. The Illumina sequencing-by-synthesis technology has been used in many studies for deep-sequencing of miRNA from different organisms to study miRNA expression profiling and discovery of new miRNAs. The throughput of a typical run from a single channel on a Solexa Genome Analyzer, for example, is ∼400–500 Mb, which includes millions of reads. Although most laboratories only perform a few runs, these laboratories are usually not equipped for large-scale computing. Some commercial packages provide a solution that clusters the tags after adaptor removal. However, further analysis on profiling, classification or distribution of small RNAs is not available. DSAP is a web server designed to provide a total solution for analyzing miRNA sequencing data generated by NGS. The functions in the DSAP suite include adaptor removal, clustering of tags and classification of ncRNAs and miRNAs based on sequencing homology using Rfam (13–16) and miRBase (17). In addition to these basic functions, DSAP also provides comparative miRNA expression profile analysis for up to five NGS datasets. These functions all together provide a global and comprehensive view on the expression profiles of miRNAs with sequence homology to known miRNAs in any organism, even those without an available reference genome.
IMPLEMENTATION
DSAP runs on a Linux CentOS 64-bit server housing two quad-core Intel® Xeon® 5300 Series Processors and 16 GB RAM installed in the Chang Gung Bioinformatics Center. Data processing is performed using Perl and Linux shell scripts. The dynamic web interface is generated using the Perl CGI library, ChartDirector for Perl and Matrix2png (18). Based on the estimation of 2 million tags per job, DSAP can handle at least 480 jobs per 24 h.
Input file and parameters
A single Solexa sequencing run produces two kinds of data. The FASTQ file contains an identifier, sequence reads and quality values for each base. The sizes of FASTQ files are usually in the gigabytes, which is not suitable for sending over the web. Another form of output format is a tab-delimited file which holds only the unique sequence read (tag) and its corresponding number of copies. A script is available from http://code.google.com/p/biopieces/wiki/read_solexa to transform the FASTQ file into unique sequence tags. The sizes of the tag files can be reduced to a few megabytes, which is more reasonable to send to a web server for analysis by web-based server tools. DSAP takes a sequence tag file as input material. After successful upload, the web server will return a page using a timestamp as an identifier to start the pipeline. The user can monitor the job status through a job status bar and several real-time bar charts recording the cleanup and clustering processes (Supplementary Figure S1a). A more detailed description of DSAP can be found on the tutorial page (http://dsap.cgu.edu.tw/tutorial.html). The only required parameter for DSAP is choosing from among 115 species, or the user can use the default of all species if the organism is not listed.
WORKFLOW
DSAP follows a series of automatic analysis steps to identify miRNAs in the input file (Figure 1): (i) cleanup to remove adaptors and poly-A/T/C/G/N nucleotides; (ii) clustering to group cleaned sequence tags into unique sequence clusters; (iii) ncRNA matching to map unique sequence clusters against the transcribed sequence library of ncRNA (Rfam); (iv) known miRNA matching to detect known miRNAs in miRBase based on sequence homology; and (v) comparative miRNAomics to show differential miRNA expression profiles from different jobs and cross-species distribution of identified miRNAs.
Figure 1.
DSAP workflow. DSAP follows several analysis steps: (a) Cleanup to remove adaptors and poly-A/T/C/G/N nucleotides. (b) Clustering to group cleaned sequence tags into unique sequence clusters. (c) ncRNA matching to map unique sequence clusters against a transcribed sequence library of ncRNA (Rfam). (d) Known miRNA matching to detect known miRNAs in miRBase based on sequence homology. (e) Comparative miRNAomics to show differential miRNA expression profiles from different jobs, and cross-species distributions of identified miRNAs.
Cleanup
To ensure the accuracy of DSAP, sequence reads that contain poly-A/T/C/G/N nucleotides or the annealing 5′-adaptor are removed. Only the sequence reads with at least 5 nt at the 3′-send matching the head of the 3′-adaptor are considered reliable reads. The user can choose whether to remove poly-A/T/C/G/N reads in the cleanup step. Sequence reads with length >16 nt after the cleanup process are retained for the clustering step. We use Supermatcher, based on the Smith–Waterman algorithm, from the EMBOSS (19–21) analysis package for the entire sequence alignment task in this step. Supermatcher combines word-match and Smith–Waterman (dynamic programming) algorithms (22). This program is more appropriate for handling a large number of sequences on web servers than using a pure dynamic programming method.
Clustering
In the clustering step, we use cleaned sequence tags from the previous step as input data to generate a set of non-redundant representative sequence clusters as output for further analysis. Sequence tags remaining after the cleanup step with 100% sequence identity and identical sequence length are grouped as non-redundant sequence clusters. Each sequence cluster has a representative Cluster ID and its total read count.
ncRNA matching
A critical step in generating small RNA libraries for NGS is the size fractionation of small RNAs from total RNA. However, in addition to miRNAs, the fractionated RNA is usually contaminated with other ncRNAs such as ribosomal RNAs (rRNAs), spliceosome RNAs (U1–U6), small nuclear RNAs (snRNAs), small nucleolar RNAs (snoRNAs) or transfer RNAs (tRNAs). Rfam, hosted by Wellcome Trust Sanger Institute in collaboration with Janelia Farm (13–16), contains information on ncRNA families. We retrieved 22 425 miRNA precursors from the Rfam database version 9.1 for use as a reference database to separate ncRNAs other than miRNAs in the non-redundant sequence tag clusters. Then, we use BLAST (with default parameters) to identify representative sequence clusters originating from rRNAs, tRNAs, snRNAs, snoRNAs or other annotated ncRNAs (23).
Known miRNA matching
miRBase is a database of published miRNA sequences and associated annotation (17). Release 14 of miRBase contains 10 883 entries representing hairpin precursor miRNAs expressing 10 581 mature miRNA products in 115 species. DSAP uses a non-redundant mature miRNA reference database created from mature miRNAs in miRBase as the default database for the identification of known miRNAs. Representative sequence clusters remaining after ncRNA matching are compared with known mature miRNA sequences with BLAST (with parameters ‘-F F -W 16’ to turn off the low complexity filter and increase the word size to 16 for increased speed). In order to obtain more reliable results, only BLAST hits with perfect alignments (100% sequence identity and cover full length of known miRNA) are retained. The hit list is summarized in a clickable bar chart that links to miRBase for further information on the identified miRNA. A tab-delimited file which contains all the details is also available for download. Representative sequence clusters that showed low sequence homology with known miRNAs are grouped as putative novel miRNAs.
Comparative miRNA analysis and cross-species distribution of miRNAs
One of the main purposes of miRNA experiments is to elucidate the differential expression levels of miRNAs among different development stages or experimental conditions. DSAP is capable of displaying non-normalized miRNA expression levels from different jobs using a log2-transformed color matrix. Furthermore, DSAP also accepts experimental results (in tab-delimited format) from other miRNA expression analyses, such as stem–loop real-time PCR, microarray or SOLiD sequencing. An example of the input file is shown in Supplementary Figure S2. Input file format details can be found on the tutorial page (http://dsap.cgu.edu.tw/tutorial.html#format).
Another powerful function of DSAP is the ability to show the distribution of identified miRNAs in different species from miRBase. This function can provide a global view on the convergence and divergence of the identified miRNAs. The users can either fill in the job identifiers provided by DSAP or to paste their own miRNA expression profiles in a text field to enable the miRNA comparison function. These functions are explained in more detail on the tutorial page (http://dsap.cgu.edu.tw/tutorial.html#miRNAomics).
RESULTS AND DISCUSSION
A working example
We provided three sequence tag files generated from small RNA libraries prepared on Day 5 (CE5), Day 7 (CE7) and Day 9 (CE9) chicken embryos (NCBI GEO database Accession Number GSE10636) as demonstration datasets (24). The user can upload a sequence tag file under 300 Mb and then choose a species or just use the default of all 115 species. The server will return a page using a timestamp as an identifier after a successful upload. Users can bookmark this page for future reference. The output page of DSAP running on the demonstration dataset is shown in Supplementary Figure S1. The output page is composed of several blocks that represent the analysis workflow of DSAP. The first block (Supplementary Figure S1a) shows the current status of the process and the time used by each step in a dynamic meter graph. The second block (Supplementary Figure S1b) shows a bar chart dynamically recording the number of sequence tags surviving the cleanup process. The third block (Supplementary Figure S1c) shows the result of clustered clean sequence tags and provides information about each unique sequence cluster in a tab-delimited file. The fourth and fifth blocks summarize the results of the unique sequence clusters matched to Rfam (Supplementary Figure S1d) and miRBase (Supplementary Figure S1e). Each matched RNA family and its related expression level is summarized in a multi-color clickable bar chart linked to miRBase for further details. All results are downloadable from the website in a tab-delimited text file. Representative sequence clusters that failed to be identified from the known miRNAs matching step can be downloaded for the identification of putative novel miRNAs. A summary of all steps in the pipeline is generated for each job (Supplementary Figure S1f). By using DSAP, 415 373 and 324 miRNAs were detected in the test datasets CE5, CE7 and CE9, respectively, out of 525 known chicken mature miRNAs deposited in miRBase. The last block (Supplementary Figure S1g) provides cross-experiments and cross-species miRNAs distribution comparison results in a color scaling matrix.
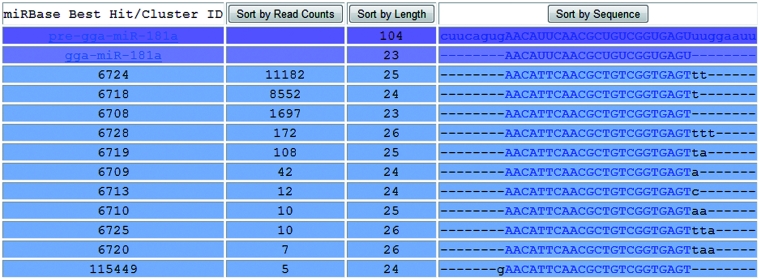
Optimized sequence alignment for isomiRs
Extensive sequence variations (isomiRs) of miRNA transcriptomes have been identified by the aid of deep-sequencing technologies (25). In addition to the detection of sequence and length variations in mature miRNAs, enzyme modification of miRNA such as RNA editing and 3′-nucleotide additions to miRNAs can also be detected by these technologies. In order to have a comprehensive view of these variations, DSAP uses a word matching method to align homologous sequences between unique sequence cluster and precursor miRNA, then append the leading and trailing sequences to obtain a multiple sequence alignment (MSA). The alignment of unique sequence clusters using our method is optimized for the observation of isomiRs and can be sorted based on expression levels of unique sequence clusters, sequence length or sequence homology (Figure 2). We found this approach to be better than using MSA methods (26–28) and more scalable in terms of computational time than local sequence clustering approaches such as CD-HIT, Uclust, BAG and BLASTclust. Because the unique sequence clusters are not equal in length, MSA algorithms attempt to make input sequences the same length by inserting gaps. In such circumstances, the leading and trailing bases of unique sequence clusters that lack homologous bases will not be aligned properly.
Figure 2.
Optimized observation of isomiRs. The alignment of unique sequence clusters with the corresponding miRNA hairpin is optimized for the observation of isomiRs. Unique sequence clusters and precursor miRNA hairpin sequences were first aligned using word matching, and then we appended the leading and trailing sequences to get a MSA. The alignment can be sorted by expression levels, sequence length or sequence homology.
Benchmarking of DSAP
We used nine NGS datasets containing 153 406–2 090 730 tags from chicken, plant and protist for benchmarking. The performance of DSAP is shown in Table 1. Most of the jobs can be completed in ∼5 min. The largest dataset, which contains over 2 million sequence tags, can be finished in 15 min.
Table 1.
Benchmarking of DSAP
| Datasets | Number of sequence tags | Processing time (hh:mm:ss) |
|---|---|---|
| CE5 | 329 334 | 00:03:42 |
| CE7 | 220 166 | 00:01:58 |
| CE9 | 153 406 | 00:01:20 |
| Protist1 | 754 059 | 00:04:38 |
| Protist2 | 736 939 | 00:05:02 |
| Protist3 | 395 939 | 00:02:50 |
| Protist4 | 697 983 | 00:03:56 |
| Plant1 | 1 643 030 | 00:11:34 |
| Plant2 | 2 090 730 | 00:13:53 |
Benchmarking is done on a Linux CentOS 64bit server housing two quad-core Intel® Xeon® 5300 Series Processors and 16 GB RAM with different numbers of sequence tags.
Comparison with other similar applications
Identification and profiling of miRNA with NGS technology is a relatively new approach. Only three other applications, miRanalyzer (29), miRExpress (30) and miRDeep (31) are available for the analysis of miRNA deep-sequencing datasets. miRanalyzer is a web server tool that performs small RNA classification and new miRNA prediction but is limited to 10 model species with the need for sequenced genomes. In addition, cross-species comparison of miRNA expression profiles is not supported. miRExpress is a stand-alone software package implemented for miRNA profiling; however, basic Linux knowledge is required to compile and execute this package. miRExpress can take deep-sequencing raw data as an input directly but lacks the ability to classify small RNAs other than miRNAs. miRDeep is also a stand-alone software package for the identification of miRNAs based on location of miRNAs on a predicted hairpin structure. Therefore, miRDeep is only useful for organisms with a known genome sequence. Compared with miRanalyzer, miRExpress and miRDeep, DSAP is the only web server tool which contains almost all of the functions of the above applications except new miRNA prediction. Furthermore, DSAP provides not only tables and text files as output formats but also clickable charts and differential miRNA expression on a color scaling matrix for better visualization. Although DSAP is not presently able to predict new miRNAs, we will add this function in our next version. Table 2 shows the key features of DSAP, miRanalyzer, miRExpress and miRDeep.
Table 2.
Comparison of DSAP, miRExpress, miRAnalyzer and miRDeep
| DSAPa | miRExpressb | miRanalyzec | miRDeepd | |
|---|---|---|---|---|
| Service type | Web server | Local server (command-line) | Web server | Local server (command-line) |
| Installation | NA | Requires knowledge on biocomputing | NA | Requires knowledge on biocomputing |
| Input file format | Tab-delimited | Tab-delimited, Fastq | Tab-delimited, Fasta | Fasta |
| miRNA database | miRBase v14 (pre-installed) | miRBase v14 | miRBase v12 (pre-installed) | miRBase v14 |
| Number of organisms available | 115 | 115 | 10 | Limited to sequenced genomes |
| Additional database | Rfam v9.1 | NA | Rfam v9.1, RepBase | UCSC Genome Browser database |
| Classification of small RNA | Yes | NA | Yes | Yes |
| Classification of miRNA | Yes | Yes | Yes | Yes |
| Iso-miRNA alignment | Yes | NA | Yes | NA |
| Cross-species distribution of miRNA | Yes (Graphic) | Yes (text) | NA | NA |
| Differential miRNA expression | Yes (Graphic) | Yes (text) | NA | NA |
| New miRNA prediction | NA | NA | Limited to 10 organisms | Limited to sequenced genomes |
| Target prediction | NA | NA | Limited to 10 organisms | Limited to sequenced genomes |
| Result output format | Table, graphic | Text file | Table | Text file |
| Result link to miRBase | Yes | NA | NA | NA |
CONCLUSION
DSAP is an ultrafast and useful tool that can process large amounts of sequencing data generated by a Solexa sequencer directly through the web and return a user-friendly report. Additionally, DSAP only takes <15 min to finish a single job of 2 million sequence tags. It is the only web-based suite designed for the identification of known miRNAs from NGS reads generated from organisms with or without a complete sequenced genome. Furthermore, DSAP also provides visualization interfaces for differential mature miRNA expression level and cross-species distribution of the identified miRNAs. A major target of DSAP in the next version will be the prediction of novel miRNAs and their putative targets.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Chang Gung Memorial Hospital (CMRPD170481, CMRPD190141 to P.T.); National Science Council, ROC (NSC97-2320-B182-011-MY3 to PT, NSC98-3112-B-007-006 to P.-C.L.); Ministry of Education, ROC to Chang Gung University and National Tsing Hua University. Funding for open access charge: NSC97-2320-B182-011-MY3.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGMENTS
We thank Dr Shu-Jen Chen and Dr Hua-Chien Chen (Department of Life Science, Chang Gung University) for their technical help and comments in preparing the manuscript.
REFERENCES
- 1.Brennecke J, Hipfner DR, Stark A, Russell RB, Cohen SM. bantam encodes a developmentally regulated microRNA that controls cell proliferation and regulates the proapoptotic gene hid in Drosophila. Cell. 2003;113:25–36. doi: 10.1016/s0092-8674(03)00231-9. [DOI] [PubMed] [Google Scholar]
- 2.Carrington JC, Ambros V. Role of microRNAs in plant and animal development. Science. 2003;301:336–338. doi: 10.1126/science.1085242. [DOI] [PubMed] [Google Scholar]
- 3.Chen CZ, Li L, Lodish HF, Bartel DP. MicroRNAs modulate hematopoietic lineage differentiation. Science. 2004;303:83–86. doi: 10.1126/science.1091903. [DOI] [PubMed] [Google Scholar]
- 4.Cheng AM, Byrom MW, Shelton J, Ford LP. Antisense inhibition of human miRNAs and indications for an involvement of miRNA in cell growth and apoptosis. Nucleic Acids Res. 2005;33:1290–1297. doi: 10.1093/nar/gki200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 6.Reinhart BJ, Slack FJ, Basson M, Pasquinelli AE, Bettinger JC, Rougvie AE, Horvitz HR, Ruvkun G. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2000;403:901–906. doi: 10.1038/35002607. [DOI] [PubMed] [Google Scholar]
- 7.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 8.Borchert GM, Lanier W, Davidson BL. RNA polymerase III transcribes human microRNAs. Nat. Struct. Mol. Biol. 2006;13:1097–1101. doi: 10.1038/nsmb1167. [DOI] [PubMed] [Google Scholar]
- 9.Du T, Zamore PD. microPrimer: the biogenesis and function of microRNA. Development. 2005;132:4645–4652. doi: 10.1242/dev.02070. [DOI] [PubMed] [Google Scholar]
- 10.Lee Y, Kim M, Han J, Yeom KH, Lee S, Baek SH, Kim VN. MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 2004;23:4051–4060. doi: 10.1038/sj.emboj.7600385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zeng Y, Yi R, Cullen BR. Recognition and cleavage of primary microRNA precursors by the nuclear processing enzyme Drosha. EMBO J. 2005;24:138–148. doi: 10.1038/sj.emboj.7600491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schwarz DS, Hutvagner G, Du T, Xu Z, Aronin N, Zamore PD. Asymmetry in the assembly of the RNAi enzyme complex. Cell. 2003;115:199–208. doi: 10.1016/s0092-8674(03)00759-1. [DOI] [PubMed] [Google Scholar]
- 13.Gardner PP, Daub J, Tate JG, Nawrocki EP, Kolbe DL, Lindgreen S, Wilkinson AC, Finn RD, Griffiths-Jones S, Eddy SR, et al. Rfam: updates to the RNA families database. Nucleic Acids Res. 2009;37:D136–D140. doi: 10.1093/nar/gkn766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Griffiths-Jones S. Annotating non-coding RNAs with Rfam. Curr. Protoc. Bioinformatics. 2005 doi: 10.1002/0471250953.bi1205s9. Chapter 12, Unit 12 15. [DOI] [PubMed] [Google Scholar]
- 15.Griffiths-Jones S, Bateman A, Marshall M, Khanna A, Eddy SR. Rfam: an RNA family database. Nucleic Acids Res. 2003;31:439–441. doi: 10.1093/nar/gkg006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Griffiths-Jones S, Moxon S, Marshall M, Khanna A, Eddy SR, Bateman A. Rfam: annotating non-coding RNAs in complete genomes. Nucleic Acids Res. 2005;33:D121–D124. doi: 10.1093/nar/gki081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Griffiths-Jones S. miRBase: the microRNA sequence database. Methods Mol. Biol. 2006;342:129–138. doi: 10.1385/1-59745-123-1:129. [DOI] [PubMed] [Google Scholar]
- 18.Pavlidis P, Noble WS. Matrix2png: a utility for visualizing matrix data. Bioinformatics. 2003;19:295–296. doi: 10.1093/bioinformatics/19.2.295. [DOI] [PubMed] [Google Scholar]
- 19.Mullan LJ, Bleasby AJ. Short EMBOSS user guide. European molecular biology open software suite. Brief. Bioinformatics. 2002;3:92–94. doi: 10.1093/bib/3.1.92. [DOI] [PubMed] [Google Scholar]
- 20.Olson SA. EMBOSS opens up sequence analysis. European molecular biology open software suite. Brief. Bioinformatics. 2002;3:87–91. doi: 10.1093/bib/3.1.87. [DOI] [PubMed] [Google Scholar]
- 21.Rice P, Longden I, Bleasby A. EMBOSS: the European molecular biology open software suite. Trends Genet. 2000;16:276–277. doi: 10.1016/s0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]
- 22.Smith TF, Waterman MS. Identification of common molecular subsequences. J. Mol. Biol. 1981;147:195–197. doi: 10.1016/0022-2836(81)90087-5. [DOI] [PubMed] [Google Scholar]
- 23.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Glazov EA, Cottee PA, Barris WC, Moore RJ, Dalrymple BP, Tizard ML. A microRNA catalog of the developing chicken embryo identified by a deep sequencing approach. Genome Res. 2008;18:957–964. doi: 10.1101/gr.074740.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kuchenbauer F, Morin RD, Argiropoulos B, Petriv OI, Griffith M, Heuser M, Yung E, Piper J, Delaney A, Prabhu AL, et al. In-depth characterization of the microRNA transcriptome in a leukemia progression model. Genome Res. 2008;18:1787–1797. doi: 10.1101/gr.077578.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Moretti S, Wilm A, Higgins DG, Xenarios I, Notredame C. R-Coffee: a web server for accurately aligning noncoding RNA sequences. Nucleic Acids Res. 2008;36:W10–W13. doi: 10.1093/nar/gkn278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thompson JD, Gibson TJ, Higgins DG. Multiple sequence alignment using ClustalW and ClustalX. Curr. Protoc. Bioinformatics. 2002 doi: 10.1002/0471250953.bi0203s00. Chapter 2, Unit 2 3. [DOI] [PubMed] [Google Scholar]
- 28.Wilm A, Higgins DG, Notredame C. R-Coffee: a method for multiple alignment of non-coding RNA. Nucleic Acids Res. 2008;36:e52. doi: 10.1093/nar/gkn174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hackenberg M, Sturm M, Langenberger D, Falcon-Perez JM, Aransay AM. miRanalyzer: a microRNA detection and analysis tool for next-generation sequencing experiments. Nucleic Acids Res. 2009;37:W68–W76. doi: 10.1093/nar/gkp347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang WC, Lin FM, Chang WC, Lin KY, Huang HD, Lin NS. miRExpress: analyzing high-throughput sequencing data for profiling microRNA expression. BMC Bioinformatics. 2009;10:328. doi: 10.1186/1471-2105-10-328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Friedlander MR, Chen W, Adamidi C, Maaskola J, Einspanier R, Knespel S, Rajewsky N. Discovering microRNAs from deep sequencing data using miRDeep. Nat. Biotechnol. 2008;26:407–415. doi: 10.1038/nbt1394. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.