Abstract
Protein–protein docking algorithms aim to predict the structure of a complex given the atomic structures of the proteins that assemble it. The docking procedure usually consists of two main steps: docking candidate generation and their refinement. The refinement stage aims to improve the accuracy of the candidate solutions and to identify near-native solutions among them. During protein–protein interaction, both side chains and backbone change their conformation. Refinement methods should model these conformational changes in order to obtain a more accurate model of the complex. Handling protein backbone flexibility is a major challenge for docking methodologies, since backbone flexibility adds a huge number of degrees of freedom to the search space. FiberDock is the first docking refinement web server, which accounts for both backbone and side-chain flexibility. Given a set of up to 100 potential docking candidates, FiberDock models the backbone and side-chain movements that occur during the interaction, refines the structures and scores them according to an energy function. The FiberDock web server is free and available with no login requirement at http://bioinfo3d.cs.tau.ac.il/FiberDock/.
INTRODUCTION
Most of the activities of living cells are performed by protein–protein interactions that form molecular complexes. Accurate modeling of the 3D structure of a complex assists in understanding its function in the cell. Additionally, atomic structures of molecular complexes are used in the field of drug design, permitting the design of small molecules that prevent or induce the formation of certain complexes. In some cases, the 3D structure of protein–protein complexes can be determined experimentally by X-ray crystallography or NMR spectroscopy. However, it is an extremely difficult and time-consuming task. Therefore, the ability to predict the structure of complexes by computational means is essential.
Protein–protein docking algorithms aim to predict the structure of a complex given the atomic structures of the proteins that assemble it. Due to protein flexibility, the structure of each individual protein (unbound conformation) is often rather different from its structure in the complex (bound conformation). Docking algorithms must therefore take the protein flexibility into account (1). This is currently the major challenge in the docking field. Protein flexibility, which includes both backbone and side-chains movements, adds a huge number of degrees of freedom to the search space, making it impossible for naïve search algorithms to find the native structure of the complex. Thus, a two-stage docking protocol is often used: performing a fast soft rigid docking (rigid docking that allows a certain amount of steric clashes), followed by flexible refinement of the results. Applying a soft rigid-docking method on the unbound structures of two proteins often results in a near-native solution that is poorly ranked due to steric clashes and bad shape complementarity. The goal of the flexible refinement stage is to model the conformational changes that the proteins undergo, and thus to resolve the clashes and improve their shape complementarity. Re-scoring the refined solutions by a binding energy score significantly improves the ranking of near-native models. Obviously, the success of the flexible refinement stage strongly depends on the existence of a near native model in the initial rigid-docking solutions.
Today, most docking refinement methods model only the side-chain flexibility and adjust the rigid-body orientations of the proteins. Modeling the backbone flexibility is considered to be a more difficult task that is addressed by only few, recently developed refinement methods (2–8).
There are many freely available web servers that deal with different aspects of the docking field. Rigid-body docking can be performed by PatchDock (9), ZDOCK (10), GRAMM-X (11), Hex (12) and SymmDock (9). ClusPro (13) filters, clusters and ranks docking solution candidates. The RosettaDock web server (14) performs local search in the vicinity of a single given input complex structure by optimizing rigid-body orientation and side-chain conformations. The NOMAD-Ref server (15) uses normal mode analysis to refine one of the molecules in a single-docking model. The FireDock web server (16), refines the rigid-body orientation and side-chain conformations of up to 1000 rigid-body solution candidates and re-scores the refined structures according to a binding energy function. The HADDOCK web server (17) performs experimental data-driven docking followed by a semi-flexible refinement.
In this article, a web server of a new flexible refinement method, called FiberDock, is presented. It is the first docking refinement web server that handles both backbone and side-chain flexibility and optimizes the relative rigid-body orientation of the proteins. Side-chain movements are modeled by a rotamer library and the backbone flexibility is modeled by an unlimited number of normal modes (18). Previous research has shown the importance of using high-frequency normal modes for modeling induced-fit conformational changes (19–21). While other, previously developed, refinement methods use only the first few normal modes, with the lowest frequency (2,3), FiberDock uses both low- and high-frequency modes. Hence, it is able to model both global and local conformational changes. The method was assessed on 20 test systems in which the backbone conformation of one protein changes upon interaction with the other. The results indicated that the incorporation of backbone flexibility in the refinement process considerably improves the accuracy and the ranking of protein complexes (21).
THE FIBERDOCK METHOD
The FiberDock method refines soft rigid-docking solution candidates and re-ranks them in order to identify the near native models (21). The refinement takes into account both backbone and side-chain flexibility. The method combines a novel normal mode analysis (NMA) based backbone refinement with our previously developed side-chain optimization and rigid-body minimization method, FireDock (22).
The NMA is performed in a pre-processing stage. In this stage, the normal modes of the proteins are calculated using the anisotropic network model (ANM) (18).
The FiberDock algorithm, which is applied on each rigid-body solution candidate, includes four main stages:
Side-chain optimization: The side-chain flexibility of interface residues of both proteins is modeled by a rotamer library. The optimal combination of rotamers is found by an integer linear programming (ILP) technique (23).
NMA-based backbone refinement: The refinement performs up to 20 iterations which consist of the following steps: (i) The van der Waals (vdW) forces that the proteins apply on each other are calculated. (ii) The 10 normal modes with the best correlation to these forces are identified, and the backbone conformation of the proteins are minimized along these normal modes. (iii) Monte Carlo (MC) rigid-body minimization is performed. (iv) A score is calculated for the current result and the result is saved if it is better than the previous results.
Rigid-body MC minimization: The rigid-body orientation of the ligand is optimized by a MC technique, and a BFGS quasi-Newton minimization is performed in each MC cycle (24,25).
Ranking according to binding energy: This stage attempts to identify near-native solutions among the entire set of refined complexes. The calculated binding energy includes a variety of energy terms, such as desolvation energy [atomic contact energy(ACE)], vdW interactions, partial electrostatics, hydrogen and disulfide bonds, π-stacking, aliphatic interactions, and more.
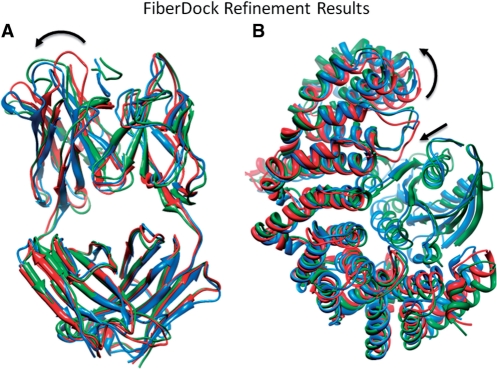
The method was tested on a set of 20 protein–protein complexes in which the receptor's interface RMSD, between its bound and the unbound conformation, varies in the range of 0.59–6.08Å. The results showed that the method successfully models backbone movements that occur during molecular interactions, and that the inclusion of the backbone refinement stage improves both the accuracy and the ranking of near-native docking solution candidates (21). Figure 1 shows the FiberDock results of refining two docking models (from our test set) that are composed of an unbound conformation of the receptor and a bound conformation of a ligand, placed in a near-native orientation. The figure shows that in both cases FiberDock correctly models the backbone movement that is essential for generating a high-accuracy docking model with no steric-clashes.
Figure 1.
FiberDock results of refining two docking models of complexes: (A) HIV-1 neutralizing antibody in complex with its V3 loop peptide antigen, PDB-ID: 1GGI and (B) Ran-Importin β, PDB-ID: 1IBR, which are composed of an unbound conformation of the receptor and a bound conformation of a ligand, placed in a near-native orientation. The unbound structure of the receptor (the starting conformation of the refinement) is colored in red and the bound complex structure is in blue. The predicted complex structure by FiberDock is in green. In both cases FiberDock correctly modeled the backbone movement (marked by arrows) that is essential for generating a high-accuracy docking model with no steric clashes. This image was produced using the UCSF Chimera package (26).
FIBERDOCK WEB-SERVER
Input
The FiberDock server can refine up to 100 rigid-docking solution candidates. The user uploads or specifies codes of two PDB (Protein Data Bank (27)) files, receptor and ligand, and provides a list of up to 100 transformations. Each transformation, when applied on the ligand, produces a candidate docking solution. If no transformation file is uploaded the identity transformation is used. Alternatively, the user can upload a PDB file that contains the rigid-docking solutions as a set of models. The candidate solutions for FiberDock can be generated by any rigid-body docking methods favored by the user (such as PatchDock (9,28), ZDOCK (10,29), GRAMM-X (11), Hex (12), etc.). In addition, the user can choose whether to model backbone movements or not. The user can also specify an e-mail address to which a link to the output web page, containing the results, will be sent when the refinement process is finished.
The server also includes optional advanced parameters for adjusting the refinement and scoring parameters for a specific biological system. These parameters are divided into four groups according to the refinement stage they affect.
For the side-chain optimization stage, the user can decide if the optimization will be preformed on both proteins, one of them or none. In addition, the user can specify the level of side-chain optimization: restricted or full. When the restricted level is chosen, only the side chains that form steric clashes will be allowed to move. The full side-chain optimization level will allow all the side chains in the protein–protein interface to be flexible. By default, the restricted level is chosen, because studies have shown that many of the side chains in the interface keep their unbound conformation within a complex (30–32).
The parameters of the backbone refinement stage include the number of lowest frequency normal modes that will be considered in the refinement. By specifying a small number (10 for example), the user restricts the backbone movements to be relatively global, whereas a high number of normal modes will allow the algorithm to use high-frequency modes, which describe local movements (if they correlate well with the chemical forces that the proteins apply on each other). In addition, the user can set the level of backbone flexibility. In order to prevent the backbone from over distorting, a penalty term is introduced into the backbone minimization step. The level of backbone flexibility determines the weight of this penalty term. The higher the level, the lower the weight. A value of 0.95 (the default value) was found to suit most of our test cases.
For the rigid-body optimization stage, the user can set the number of MC iterations. In general, increasing this value improves the search for a local minima in the vicinity of the ligand's current position. However, according to our experience, the optimization usually converges after 50 iterations.
The complex type parameter (Default, Antibody-Antigen or Enzyme-Inhibitor), is used for adjusting the weights of the scoring function for a specific biological system. The parameter of atomic radius scale influences the extent of acceptable steric clashes in the final refined solutions. This parameter scales down the radius of the atoms, affecting the VdW terms that are used in all of the three refinement stages and the final calculated binding energy.
Output
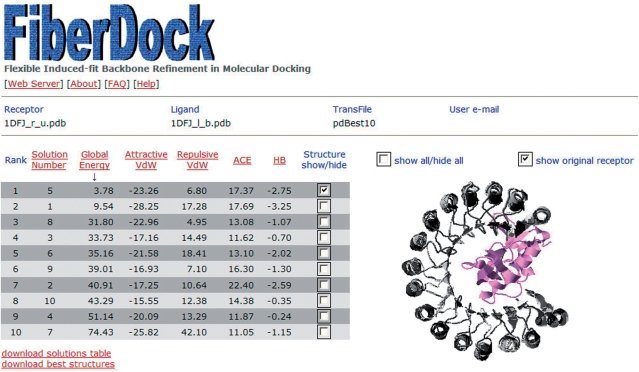
When the refinement is finished, a web page with the results is generated and a link to it is sent to the e-mail address specified by the user. This web page (Figure 2) contains a table in which each row corresponds to a single refined solution. Each row specifies the rank of the solution according to the binding energy value, its original number (according to the given transformation file), the global binding energy value and the values of four of the energy terms (Attractive VdW, repulsive VdW, ACE and hydrogen bonds). The table is sorted by the binding energy of the refined solution. The user can view the 3D structure of each refined complex in a Jmol applet window (33). The different structures can be viewed simultaneously, allowing the user to easily compare different models. The PDB files of the refined solutions can be downloaded, and so can the full results table that details the values of all the energy terms, for each solution. This table also specifies the linear combination of normal modes that generates the refined backbone conformation of the receptor and the ligand.
Figure 2.
The output of FiberDock web server. The results are presented in a table, sorted by the binding energy value. The user can view the 3D structure of each of the refined complex in a Jmol applet window.
CONCLUSIONS
Handling backbone flexibility is currently the main challenge in the docking field. In many cases, even a slight backbone movement prevents near-native rigid-docking solutions from being highly ranked, since these models will often contain steric clashes. Therefore, flexible refinement is needed in order to resolve these clashes by backbone and side-chain movements and a minimization of the rigid-body orientation. The FiberDock method was developed to meet this challenge. This new method mimics an induced fit-process. The backbone and side-chain movements are inferred from the vdW forces that the proteins apply on each other. The method models backbone movements by normal modes. It uses both low- and high-frequency modes and therefore is able to model both global and local conformational changes, such as opening of binding sites and loop movements.
In order to make this method available for the entire biological community, a clear and user-friendly web server was developed, which requires no previous knowledge in docking algorithms. This is the first web server for flexible docking refinement, which models both backbone and side-chain flexibility. It refines a single rigid-body docking solution in an average time of 14s. Therefore, it can be used for refining and re-ranking of up to 100 solutions in a reasonable time. The FiberDock software (for Linux users) can also be downloaded from the web site. The downloaded version does not restrict the amount of refined docking solutions. We believe that this server will be very useful to the biological community. It can help model new structures of protein–protein complexes and as such improve our understanding of protein functions in the living cell.
FUNDING
Adams fellowship of the Israel Academy of Sciences and Humanities (to E.M., in part); Israel Science Foundation (grant no. 1403/09, to H.J.W., in part); Hermann Minkowski Minerva Geometry Center; National Cancer Institute, National Institutes of Health (Federal funds, contract number HHSN261200800001E). Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research (in part). Funding for open access charge: National Institutes of Health (contract number HHSN261200800001E).
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
Molecular graphics images were produced using the UCSF Chimera package from the Resource for Biocomputing, Visualization, and Informatics at the University of California, San Francisco. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the US Government.
REFERENCES
- 1.Andrusier N, Mashiach E, Nussinov R, Wolfson HJ. Principles of flexible protein-protein docking. Proteins. 2008;73:271–289. doi: 10.1002/prot.22170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lindahl E, Delarue M. Refinement of docked protein-ligand and protein-DNA structures using low frequency normal mode amplitude optimization. Nucleic Acids Res. 2005;33:4496–4506. doi: 10.1093/nar/gki730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.May A, Zacharias M. Energy minimization in low-frequency normal modes to efficiently allow for global exibility during systematic protein-protein docking. Proteins. 2008;70:794–809. doi: 10.1002/prot.21579. [DOI] [PubMed] [Google Scholar]
- 4.May A, Zacharias M. Protein-protein docking in CAPRI using ATTRACT to account for global and local flexibility. Proteins. 2007;69:774–780. doi: 10.1002/prot.21735. [DOI] [PubMed] [Google Scholar]
- 5.Wang C, Bradley P, Baker D. Protein-protein docking with backbone flexibility. J. Mol. Biol. 2007;373:503–519. doi: 10.1016/j.jmb.2007.07.050. [DOI] [PubMed] [Google Scholar]
- 6.Chaudhury S, Sircar A, Sivasubramanian A, Berrondo M, Gray JJ. Incorporating biochemical information and backbone flexibility in RosettaDock for CAPRI rounds 6-12. Proteins. 2007;69:793–800. doi: 10.1002/prot.21731. [DOI] [PubMed] [Google Scholar]
- 7.Fitzjohn PW, Bates PA. Guided docking: first step to locate potential binding sites. Proteins. 2003;52:28–32. doi: 10.1002/prot.10380. [DOI] [PubMed] [Google Scholar]
- 8.Król M, Chaleil RA, Tournier AL, Bates PA. Implicit flexibility in protein docking: cross-docking and local refinement. Proteins. 2007;69:750–757. doi: 10.1002/prot.21698. [DOI] [PubMed] [Google Scholar]
- 9.Schneidman-Duhovny D, Inbar Y, Nussinov R, Wolfson HJ. PatchDock and SymmDock: servers for rigid and symmetric docking. Nucleic Acids Res. 2005;33:W363–W367. doi: 10.1093/nar/gki481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen R, Li L, Weng Z. ZDOCK: an initial-stage protein-docking algorithm. Proteins. 2003;52:80–87. doi: 10.1002/prot.10389. [DOI] [PubMed] [Google Scholar]
- 11.Tovchigrechko A, Vakser IA. GRAMM-X public web server for protein–protein docking. Nucleic Acids Res. 2006;34:310–314. doi: 10.1093/nar/gkl206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ritchie DW, Kemp G.JL. Protein docking using spherical polar Fourier correlations. Proteins. 2000;39:178–194. [PubMed] [Google Scholar]
- 13.Comeau SR, Gatchell DW, Vajda S, Camacho CJ. Cluspro: a fully automated algorithm for protein-protein docking. Nucleic Acids Res. 2004;32:W96–W99. doi: 10.1093/nar/gkh354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lyskov S, Gray JJ. The RosettaDock server for local protein-protein docking. Nucleic Acids Res. 2008;36:W233–W238. doi: 10.1093/nar/gkn216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lindahl E, Azuara C, Koehl P, Delarue M. NOMAD-Ref: visualization, deformation and refinement of macromolecular structures based on all-atom normal mode analysis. Nucleic Acids Res. 2006;34:W52–W56. doi: 10.1093/nar/gkl082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mashiach E, Schneidman-Duhovny D, Andrusier N, Nussinov R, Wolfson HJ. FireDock: a web server for fast interaction refinement in molecular docking. Nucleic Acids Res. 2008;36:W229–W232. doi: 10.1093/nar/gkn186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dominguez C, Boelens R, Bonvin A. HADDOCK: a protein-protein docking approach based on biochemical or biophysical information. J. Am. Chem. Soc. 2003;125:1731–1737. doi: 10.1021/ja026939x. [DOI] [PubMed] [Google Scholar]
- 18.Hinsen K. Analysis of domain motions by approximate normal mode calculations. Proteins. 1998;33:417–429. doi: 10.1002/(sici)1097-0134(19981115)33:3<417::aid-prot10>3.0.co;2-8. [DOI] [PubMed] [Google Scholar]
- 19.Petrone P, Pande VS. Can conformational change be described by only a few normal modes? Biophys. J. 2006;90:1583–1593. doi: 10.1529/biophysj.105.070045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cavasotto CN, Kovacs JA, Abagyan RA. Representing receptor flexibility in ligand docking through relevant normal modes. J. Am. Chem. Soc. 2005;127:9632–9640. doi: 10.1021/ja042260c. [DOI] [PubMed] [Google Scholar]
- 21.Mashiach E, Nussinov R, Wolfson HJ. FiberDock: flexible induced-fit backbone refinement in molecular docking. Proteins. 2009;78:1503–1519. doi: 10.1002/prot.22668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Andrusier N, Nussinov R, Wolfson HJ. FireDock: fast interaction refinement in molecular docking. Proteins. 2007;69:139–159. doi: 10.1002/prot.21495. [DOI] [PubMed] [Google Scholar]
- 23.Eriksson O. Side chain-positioning as an integer programming problem. Lect. Notes Comput. Sci. 2001;2149:128–141. [Google Scholar]
- 24.Broyden CG. The convergence of a class of double-rank minimization algorithms. J. Inst. Math. Appl. 1970;6:76–90. [Google Scholar]
- 25.Fletcher R. A new approach to variable metric algorithms. The Computer Journal. 1970;13:317–322. [Google Scholar]
- 26.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera - a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 27.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The protein data bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Duhovny D, Nussinov R, Wolfson HJ. Efficient unbound docking of rigid molecules. Lect. Notes Comput. Sci. 2002;2452:185–200. [Google Scholar]
- 29.Chen R, Weng Z. Docking unbound proteins using shape complementarity, desolvation, and electrostatics. Proteins. 2002;47:281–294. doi: 10.1002/prot.10092. [DOI] [PubMed] [Google Scholar]
- 30.Smith GR, Sternberg M.JE, Bates PA. The relationship between the flexibility of proteins and their conformational states on forming protein-protein complexes with application to protein-protein docking. J. Mol. Biol. 2005;347:1077–1101. doi: 10.1016/j.jmb.2005.01.058. [DOI] [PubMed] [Google Scholar]
- 31.Rajamani D, Thiel S, Vajda S, Camacho CJ. Anchor residues in protein-protein interactions. Proc. Natl Acad. Sci. USA. 2004;101:11287–11292. doi: 10.1073/pnas.0401942101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li X, Keskin O, Ma B, Nussinov R, Liang J. Protein-protein interactions: hot spots and structurally conserved residues often locate in complemented pockets that pre-organized in the unbound states: implications for docking. J. Mol. Biol. 2004;344:781–795. doi: 10.1016/j.jmb.2004.09.051. [DOI] [PubMed] [Google Scholar]
- 33.Herraez A. Biomolecules in the computer: Jmol to the rescue. Biochem. Mol. Biol. Educ. 2006;34:255–261. doi: 10.1002/bmb.2006.494034042644. [DOI] [PubMed] [Google Scholar]