Abstract
We present TranslatorX, a web server designed to align protein-coding nucleotide sequences based on their corresponding amino acid translations. Many comparisons between biological sequences (nucleic acids and proteins) involve the construction of multiple alignments. Alignments represent a statement regarding the homology between individual nucleotides or amino acids within homologous genes. As protein-coding DNA sequences evolve as triplets of nucleotides (codons) and it is known that sequence similarity degrades more rapidly at the DNA than at the amino acid level, alignments are generally more accurate when based on amino acids than on their corresponding nucleotides. TranslatorX novelties include: (i) use of all documented genetic codes and the possibility of assigning different genetic codes for each sequence; (ii) a battery of different multiple alignment programs; (iii) translation of ambiguous codons when possible; (iv) an innovative criterion to clean nucleotide alignments with GBlocks based on protein information; and (v) a rich output, including Jalview-powered graphical visualization of the alignments, codon-based alignments coloured according to the corresponding amino acids, measures of compositional bias and first, second and third codon position specific alignments. The TranslatorX server is freely available at http://translatorx.co.uk.
INTRODUCTION
Evolutionary comparisons of primary sequence data rely on the generation of a multiple sequence alignment that maximizes the likelihood of positional homology between nucleotides or amino acids by introducing gaps (1). During the course of evolution, functional and structural constraints leave their footprint on sequences in the form of mutations, insertions and deletions. Different regions of a molecule, depending on their functional or structural importance, are subject to different selective forces, which result in evolutionary rate heterogeneity (2). In those regions that are well-conserved, saturation is low, indels are rare and assigning positional homology is straightforward. However, in those regions experiencing faster substitution rates and more frequent indels, assessing positional homology is more problematic.
Available methods for producing multiple alignments of nucleic acids do not take account of the important fact that coding DNA evolves as triplets of nucleotides or codons; insertions and deletions in coding genes are expected to occur in sets of three nucleotides to avoid altering the coding reading frame. Alignments of coding DNA can also be improved by a consideration of the amino acid sequences that the DNA codes for—this is because amino acid sequences change more slowly than their nucleic acid counterparts and are therefore easier to align. The greater rapidity of change and corresponding difficulty aligning nucleic acids results principally from the degeneracy of the genetic code sequence—synonymous nucleotide changes are not selected against. Added to this, the larger amino acid alphabet (20 amino acids versus 4 nucleotides) results in a higher likelihood of homoplasy (e.g. convergent evolution) between DNA sequences compared to amino acid sequences. Finally, the frequent substitutions that occur between physico-chemically similar amino acids are easily accounted for when aligning, yet these changes lead to less easily modelled substitutions in DNA.
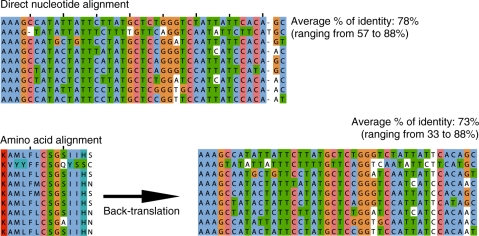
Due to the different evolutionary behaviour of nucleotide and amino acid sequences, and because selection acts most prominently at the protein level, amino acid translations of two orthologous protein-coding genes can share a higher percentage of identity than the corresponding nucleotide versions even if their alphabet is five times larger. Consequently, evolutionary differences between DNA and protein languages make the alignment of divergent nucleotide sequences considerably more difficult than of their corresponding amino acid translations. A straightforward approach to circumvent this limitation and to align nucleotide sequences accurately is to translate the DNA sequences into amino acids, align these amino acids and then back-translate the alignment to the nucleotide alphabet. Figure 1 shows a real example from a region of the ND5 mitochondrial gene, in which the limitations of the direct nucleotide alignment are manifest and avoided with the back-translation approach.
Figure 1.
Example illustrating the different performance of the direct and back-translated nucleotide alignments (multiple alignments were built with Muscle with default parameters).
Several tools have been developed based on this principle of back-translation, including stand-alone programs such as transAlign (3), protal2dna (4), and tranalign (5), as well as web servers including RevTrans (6), PROTOGENE (7) and PAL2NAL (8); each of these tools has different limitations. All these solutions, with the exception of RevTrans, require that the user provide the amino acid alignment together with nucleotide sequences, some of the packages do not provide a complete list of available genetic codes (PAL2NAL) and, except for protal2dna, none of these tools allows the user to assign a different genetic code to each of the nucleotide sequences. Finally, only PAL2NAL and transAlign are designed to consider cases in which frame shifts occur (as e.g. in the case of pseudogenes).
Here, we present a new web server, TranslatorX, built on this principle of using the translated amino acid alignment to guide the alignment of nucleotide sequences. The new program is designed to avoid the limitations exhibited by previous related tools. TranslatorX offers a battery of different multiple alignment programs, automatic translation based on all documented genetic codes (more than one of which can be used simultaneously), automatic identification of the coding reading frame and nucleotide codon disambiguation according to IUPAC nomenclature (9). In addition, TranslatorX provides an information-rich output aimed at guiding subsequent analyses based on the resulting multiple alignments—typically phylogenetic reconstruction or calculation of synonymous versus non-synonymous substitution rates.
TRANSLATORX SERVER
TranslatorX usage is simple and highly customizable. The basic usage requires a set of nucleotide sequences as input. Most sequence formats are supported, thanks to the ReadSeq sequence format conversion tool (10). By default, all sequences are translated according to the standard/universal nuclear genetic code. Alternative codes can be specified, either as a single alternative for all sequences in the alignment or as specific variants for each of the sequences.
Assigning multiple different genetic codes can be accomplished either through interactive menus that help the user to define the code of each species, or using a predefined text file that can be copied and pasted or uploaded from file. The format used to define the genetic code for each species is the name of the taxon (or sequence) plus the index of the corresponding genetic code separated by a tab or comma (e.g. Bolinus brandaris, 4). All documented genetic codes are incorporated in TranslatorX; the list includes those codes defined in NCBI and GenBank plus two additional ones: the ancestral Arthropod mitochondrial genetic code (11) and the Hemichordate mitochondrial code (our unpublished data). Several different multiple alignment programs including Muscle (12), Mafft (13), T-Coffee (14), Prank (15) and ClustalW (16) can be chosen to align the amino acids. As an alternative, users are able to upload their own pre-calculated protein alignment.
By default, TranslatorX expects the input nucleotide sequences to be in frame +1, however, if multiple stop codons suggest that this is not the case, a warning is given and the server may be requested to determine the most likely coding frame automatically. This is done on the basis that the reading frame with the fewest stop codons (ideally none) is the most likely coding frame. In contrast to PAL2NAL and transAlign (3,8), frameshifts cannot be accommodated.
Removal of ambiguously aligned regions is a common practice in phylogenetic studies. Programs such as GBlocks (17) are designed to identify and remove highly variable regions of the alignment where positional homology is dubious. TranslatorX provides an innovative approach in the process of alignment cleaning that minimizes deleted regions: rather than cleaning the nucleotide alignment based on its intrinsic positional information, TranslatorX uses GBlocks to analyse the amino acid alignment and removes columns from the nucleotide alignment based on this analysis. The resulting nucleotide alignment may retain highly variable (and informative) regions, but the user can be confident about their positional homology.
TRANSLATORX OUTPUT
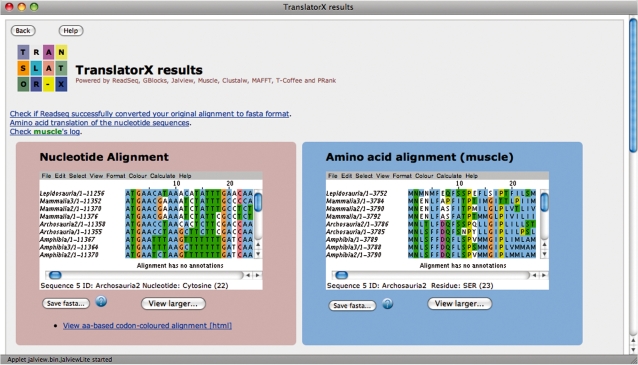
The principal outputs of the program are alignments visualized within a Jalview window (18). Three outputs are presented: amino acids (aa), nucleotides (nt) and the same nucleotide alignment coloured according to the amino acids coded by each triplet. The Jalview viewer has additional interesting features incorporated such as the ability to reconstruct neighbour-joining trees or to refine the alignment by manual editing (Figure 2). If the user has selected the alignment cleaning option, two additional alignments will be displayed: the GBlocks-cleaned amino acid alignment and the corresponding cleaned nucleotide alignment.
Figure 2.
Screen capture of a fragment of the results of TranslatorX. The nucleotide back-translated alignment and the corresponding amino acid alignment are shown with Jalview.
The output of TranslatorX is particularly aimed to be useful for downstream phylogenetic analyses. For this reason, the final nucleotide alignments (either the complete or the cleaned alignment) are available divided into different alignments derived from different subsets of the three codon positions—third codon positions are often left out of phylogenetic analyses due to substitutional saturation. The first, second, third and first + second codon positions alignments can be displayed and downloaded individually.
Additionally, the nucleotide compositional bias is calculated for each sequence and for each codon position—such biases can cause systematic errors in phylogenetic analyses and use of these options can indicate if cleaning the alignment reduces biases, or if a particular sequence or codon position is strongly biased.
COMPARISON OF THE TRANSLATORX AND DIRECT NUCLEOTIDE ALIGNMENTS IN TERMS OF PHYLOGENETIC INFERENCE PERFORMANCE
In order to illustrate the benefits of using TranslatorX, we analysed a concatenated data set of 13 mitochondrial protein-coding genes from nine vertebrate species covering a diverse range of sequence similarity, including the following taxonomic groups: Euarchontoglires and Laurasiatheria (both placental mammals), Metatheria (marsupials), Monotremata (platypus), Testudines (turtles), Squamata (lizards and snakes), Amphibia (frogs, salamanders and caecilians) and Coelacanthimorpha (coelacanths). One species of Actinopterygii (ray-finned fishes) was used as an outgroup.
We aligned nucleotide sequences directly with Mafft, Muscle, ClustalW and T-Coffee. In addition, we built back-translated nucleotide alignments using TranslatorX with the amino acid alignment step performed using the same alignment programs. We compared each possible pair among the eight resulting alignments and determined how many positions varied between the two alignments compared. The results revealed a lower variance between the TranslatorX alignments (Table 1), suggesting that using amino acid information results in a better alignment performance. In addition, the number of gap segments (equivalent to gap openings) and total number of gaps were lower in back-translated alignments than in direct nucleotide alignments (Table 2). Next, we tested the reliability of the different alignments by analysing their phylogenetic performance. Phylogenetic trees were inferred using the maximum likelihood-based software Phyml v3.0 (19) using the best-fit model GTR+I+G as identified by ModelTest (20). In spite of the differences between alignments, the topology of tree recovered was stable.
Table 1.
Length of each alignment and number of positions whose alignments differ between each pair of methods
| Length | TrX + ClustalW | TrX + Muscle | TrX + Mafft | TrX + Tcoffee | ClustalW | Muscle | Mafft | Tcoffee | |
|---|---|---|---|---|---|---|---|---|---|
| TrX + ClustalW | 11 514 | 0 | 780 | 816 | 684 | 1580 | 1260 | 1491 | 2520 |
| TrX + Muscle | 11 553 | 819 | 0 | 501 | 633 | 1582 | 1246 | 1455 | 2543 |
| TrX + Mafft | 11 562 | 864 | 510 | 0 | 693 | 1545 | 1240 | 1448 | 2520 |
| TrX + Tcoffee | 11 526 | 696 | 606 | 657 | 0 | 1552 | 1201 | 1450 | 2505 |
| ClustalW | 11 562 | 1628 | 1591 | 1545 | 1588 | 0 | 1338 | 1380 | 2434 |
| Muscle | 11 604 | 1350 | 1297 | 1282 | 1279 | 1380 | 0 | 1342 | 2487 |
| Mafft | 11 679 | 1656 | 1581 | 1565 | 1603 | 1497 | 1417 | 0 | 2574 |
| Tcoffee | 13 771 | 4777 | 4761 | 4729 | 4750 | 4643 | 4654 | 4666 | 0 |
Trx, TranslatorX—the back-translation approach.
Table 2.
Number of gaps, gap segments and types of gap arrangements for the different alignments
| ClustalW | Muscle | Mafft | T-coffee | SD | |
| Alignment length | 11 562 | 11 604 | 11 679 | 13771 | 1079.09 |
| Total gaps | 1803 | 2181 | 2856 | 21 684 | 9711.77 |
| Gap segments | 536 | 407 | 431 | 2414 | 979.60 |
| One gap | 236 | 133 | 94 | 1179 | 515.82 |
| Two gaps | 146 | 94 | 82 | 579 | 237.46 |
| Three gaps | 425 | 620 | 866 | 6449 | 2911.60 |
| TrX + ClustalW | TrX + Muscle | TrX + Mafft | TrX + Tcoffee | SD | |
| Alignment length | 11 514 | 11 553 | 11 562 | 11 526 | 22.50 |
| Total gaps | 1371 | 1722 | 1803 | 1479 | 202.50 |
| Gap segments | 166 | 213 | 232 | 206 | 27.77 |
| One gap | 0 | 0 | 0 | 0 | 0.00 |
| Two gaps | 0 | 0 | 0 | 0 | 0.00 |
| Three gaps | 457 | 574 | 601 | 493 | 67.50 |
Trx, TranslatorX—the back-translation approach.
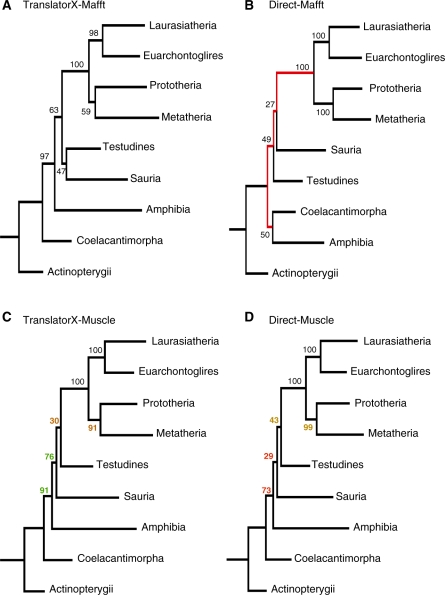
To measure the benefits of TranslatorX further, for each alignment method, we extracted those positions whose alignment differed between the TranslatorX alignment (back-translated) and the direct alignments; these sub-alignments contain those regions that are most variable and hence most difficult to align. Each pair of these variable sub-alignments was used independently to reconstruct the phylogeny of these taxonomic groups. We compared each pair of trees to an optimal reference tree obtained with the portion of data that did not vary between TranslatorX and direct alignment methods. We found that, in the majority of cases, the back-translated subalignments produced trees more similar to the reference tree. In the case of Mafft, the improvement was visible directly in the tree topology (Figure 3A–B). In the case of Muscle, the topology did not vary, but two tree nodes had a higher bootstrap support with the TranslatorX alignment (Figure 3C–D); these two nodes correspond to expected clades (according to current knowledge). In contrast, two nodes obtained a lower bootstrap support with the TranslatorX method, and in both cases the nodes with reduced support correspond to clades of questionable validity: Sauria + (Testudines + Mammals) and (Metatheria + Monotremata) + Placentals. The phylogenetic trees obtained with the ClustalW and T-Coffee alignments are provided as Supplementary Data. These results conform with the expectation that the use of amino acids maximizes the correct interpretation of positional homology in variable regions resulting in better phylogenetic performance (higher support for good nodes and lower support for bad nodes).
Figure 3.
Comparison of the phylogenetic trees inferred from the sub-alignments that comprise positions whose alignment differed between the back-translated and direct Mafft (A, B) and Muscle (C, D) alignments.
With respect to the nucleotide compositional bias, TranslatorX calculated an overall GC content of 40.36% (ranging from 36.12% to 45.30% for different species). The observed bias was slightly but significantly (P-value <0.001) reduced for all taxa after alignment cleaning (490 positions out of 3851 were discarded from the amino acid alignment), yielding an overall GC content of 40.83% (Table 3). The GC content of the discarded positions was more biased (38.08%) indicating that the characteristic bias of metazoan mitochondrial genomes accumulates more strongly in variable regions (and third codon positions). When we compared the GC content of the alignment regions that varied between TranslatorX alignment and direct alignment against the GC content of the regions that did not vary, we detected that the bias accumulated more strongly in the former (37.47% versus 40.68%). With respect to the three codon positions, the bias was most significantly reduced for first positions after GBlocks cleaning. Interestingly, third positions, which initially display the most bias, were not significantly affected by the cleaning. GBlocks cleaning also increased the percentage of sequence identity and eliminated all gaps. A similar analysis was conducted for the subalignments obtained after comparing the TranslatorX and direct approaches (Table 3). As expected, the differing sub-alignments are particularly variable and rich in gaps, and also encompass a particularly biased nucleotide composition compared to the positions whose alignment did not vary between the two approaches. Interestingly, the average percentage identity was greater for the direct sub-alignment than for the TranslatorX one.
Table 3.
Statistics for the different alignments obtained using the Muscle alignment program
| Length | Average %id | Min %id | Max %id | GC (%) | Gaps (%) | |
|---|---|---|---|---|---|---|
| TranslatorX | ||||||
| Complete alignment | 11 553 | 67 | 63 | 71 | 40.36 | 1.65 |
| Gblocks accepted | 10 083 | 69 | 66 | 73 | 40.83 | 0 |
| Gblocks discarded | 1470 | 46 | 39 | 57 | 36.67 | 13.02 |
| TranslatorX versus direct Muscle | ||||||
| Consensus (coinciding) | 10 237 | 69 | 65 | 73 | 40.68 | 0.06 |
| Different in TranslatorX | 1316 | 45 | 36 | 58 | 37.47 | 14.08 |
| Different in Muscle | 1367 | 49 | 43 | 59 | 37.47 | 17.29 |
The first set of rows are comparisons of TranslatorX alignments before and after the GBlocks cleaning. The second set of rows refers to the comparison between TranslatorX and direct nucleotide alignment approaches. Average %id, Min %id and Max %id: average/minimum/maximum percentage of identity between aligned sequences; GC %: GC-content percentage; Gaps %: percentage of gaps in the multiple alignment.
DISCUSSION
Positional homology is best established at the amino acid level due to the evolution of coding DNA as triplets of nucleotides, the degeneracy of the genetic code and the larger alphabet of proteins that slow sequence similarity degradation and saturation phenomena. Logic suggests that amino acid alignment information can be used to obtain a more reliable nucleotide sequence alignment. We have developed a web-based tool (TranslatorX) that generates back-translated alignments and compared their phylogenetic performance with respect to direct nucleotide alignments in recovering the phylogenetic relationships of a set of vertebrates. We find that, even when the nucleotide sequences were closely related and showed high similarity, important differences between the back-translated and direct approaches were seen at several levels: (i) alignment concordance between different aligning methods; (ii) the number and arrangement of gaps; and (iii) the evolutionary information content in the most variable regions.
Our web server also provides an innovative approach to clean nucleotide alignments based on initial GBlocks cleaning of the corresponding amino acid alignment. As a result of using TranslatorX, the user can be more confident that variable positions in the back-translated alignment are properly aligned and positional homology is ensured. This approach also maximizes the retention of variable positions that otherwise would be removed by GBlocks cleaning of the nucleotide alignment.
Our results also show that nucleotide compositional biases in this data set are concentrated in variable regions; as represented either by the regions removed by GBlocks or in the sites differently aligned by the two alternative approaches. The bias on first codon positions was most significantly affected by the cleaning. Second codon position bias was affected to a lesser extent, probably because second positions are the least variable. Unexpectedly, third codon positions, the most variable and most biased, were not affected by alignment cleaning. This might be due to the fact that synonymous changes are frequent, and mutational saturation might have been reached both in variable and conserved regions of the alignment.
The comparison of the direct and back-translated nucleotide alignments reveals that the direct approach usually results in alignments with higher percentages of identities and these alignments look better on casual inspection. Our results indicate, however, that the increase in the percentage of identities is reached by introducing many more gaps and by misaligning homologous sites. This does not reflect a problem in the multiple alignment programs (in fact, the alignment score is higher on direct than on back-translated alignments). Instead, the limitations of the direct alignment of nucleotide sequences arise from ignoring biological forces that affect coding DNA.
There is an open debate regarding whether nucleotide or amino acid characters should be preferred for phylogenetic inference (21). Some authors argue that slowly evolving characters (e.g. amino acids) are preferable to fast evolving ones (nucleotides) for inferring deep evolutionary relationships. Other authors have found that nucleotides, even if more saturated, might encompass a better phylogenetic signal (21–23). Although there is no consensus regarding this dilemma, it is clear that the alignment of nucleotide sequences is best accomplished when protein information is taken into account.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
Conflict of interest statement. None declared.
REFERENCES
- 1.Talavera G, Castresana J. Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst. Biol. 2007;56:564–577. doi: 10.1080/10635150701472164. [DOI] [PubMed] [Google Scholar]
- 2.Yang Z. Maximum-likelihood estimation of phylogeny from DNA sequences when substitution rates differ over sites. Mol. Biol. Evol. 1993;10:1396–1401. doi: 10.1093/oxfordjournals.molbev.a040082. [DOI] [PubMed] [Google Scholar]
- 3.Bininda-Emonds OR. transAlign: using amino acids to facilitate the multiple alignment of protein-coding DNA sequences. BMC Bioinformatics. 2005;6:156. doi: 10.1186/1471-2105-6-156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schuerer K, Letondal C. 2003. [(26 April, 2010, date last accessed)]. PROTAL2DNA: align DNA sequences given the corresponding protein alignment. http://www.pasteur.fr/recherche/unites/sis/formation/bioperl/lecture_code/protal2dna.html. [Google Scholar]
- 5.Rice P, Longden I, Bleasby A. EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet. 2000;16:276–277. doi: 10.1016/s0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]
- 6.Wernersson R, Pedersen AG. RevTrans: multiple alignment of coding DNA from aligned amino acid sequences. Nucleic Acids Res. 2003;31:3537–3539. doi: 10.1093/nar/gkg609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Moretti S, Reinier F, Poirot O, Armougom F, Audic S, Keduas V, Notredame C. PROTOGENE: turning amino acid alignments into bona fide CDS nucleotide alignments. Nucleic Acids Res. 2006;34:W600–603. doi: 10.1093/nar/gkl170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Suyama M, Torrents D, Bork P. PAL2NAL: robust conversion of protein sequence alignments into the corresponding codon alignments. Nucleic Acids Res. 2006;34:W609–612. doi: 10.1093/nar/gkl315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Panico R, Powell WH, Riche JC. A guide to IUPAC Nomenclature of Organic Compounds. Oxford, UK: Blackwell Scientific Publications; 1993. [Google Scholar]
- 10.Gilbert D. 2001. ReadSeq: read & reformat biosequences. http://iubio.bio.indiana.edu/ [Google Scholar]
- 11.Abascal F, Posada D, Knight RD, Zardoya R. Parallel evolution of the genetic code in arthropod mitochondrial genomes. PLoS Biol. 2006;4:e127. doi: 10.1371/journal.pbio.0040127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Katoh K, Kuma K, Toh H, Miyata T. MAFFT version 5: improvement in accuracy of multiple sequence alignment. Nucleic Acids Res. 2005;33:511–518. doi: 10.1093/nar/gki198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Notredame C, Higgins DG, Heringa J. T-Coffee: a novel method for fast and accurate multiple sequence alignment. J. Mol. Biol. 2000;302:205–217. doi: 10.1006/jmbi.2000.4042. [DOI] [PubMed] [Google Scholar]
- 15.Loytynoja A, Goldman N. An algorithm for progressive multiple alignment of sequences with insertions. Proc. Natl Acad. Sci. USA. 2005;102:10557–10562. doi: 10.1073/pnas.0409137102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Castresana J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol. Biol. Evol. 2000;17:540–552. doi: 10.1093/oxfordjournals.molbev.a026334. [DOI] [PubMed] [Google Scholar]
- 18.Clamp M, Cuff J, Searle SM, Barton GJ. The Jalview Java alignment editor. Bioinformatics. 2004;20:426–427. doi: 10.1093/bioinformatics/btg430. [DOI] [PubMed] [Google Scholar]
- 19.Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- 20.Posada D, Crandall KA. MODELTEST: testing the model of DNA substitution. Bioinformatics. 1998;14:817–818. doi: 10.1093/bioinformatics/14.9.817. [DOI] [PubMed] [Google Scholar]
- 21.Simmons MP, Carr TG, O’Neill K. Relative character-state space, amount of potential phylogenetic information, and heterogeneity of nucleotide and amino acid characters. Mol. Phylogenet Evol. 2004;32:913–926. doi: 10.1016/j.ympev.2004.04.011. [DOI] [PubMed] [Google Scholar]
- 22.Townsend JP, Lopez-Giraldez F, Friedman R. The phylogenetic informativeness of nucleotide and amino acid sequences for reconstructing the vertebrate tree. J. Mol. Evol. 2008;67:437–447. doi: 10.1007/s00239-008-9142-0. [DOI] [PubMed] [Google Scholar]
- 23.Simmons MP, Ochoterena H, Freudenstein JV. Amino acid vs. nucleotide characters: challenging preconceived notions. Mol. Phylogenet Evol. 2002;24:78–90. doi: 10.1016/s1055-7903(02)00202-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.