Abstract
The proteome-wide characterization and analysis of protein ligand-binding sites and their interactions with ligands can provide pivotal information in understanding the structure, function and evolution of proteins and for designing safe and efficient therapeutics. The SMAP web service (SMAP-WS) meets this need through parallel computations designed for 3D ligand-binding site comparison and similarity searching on a structural proteome scale. SMAP-WS implements a shape descriptor (the Geometric Potential) that characterizes both local and global topological properties of the protein structure and which can be used to predict the likely ligand-binding pocket [Xie,L. and Bourne,P.E. (2007) A robust and efficient algorithm for the shape description of protein structures and its application in predicting ligand-binding sites. BMC bioinformatics, 8 (Suppl. 4.), S9.]. Subsequently a sequence order independent profile–profile alignment (SOIPPA) algorithm is used to detect and align similar pockets thereby finding protein functional and evolutionary relationships across fold space [Xie, L. and Bourne, P.E. (2008) Detecting evolutionary relationships across existing fold space, using sequence order-independent profile-profile alignments. Proc. Natl Acad. Sci. USA, 105, 5441–5446]. An extreme value distribution model estimates the statistical significance of the match [Xie, L., Xie, L. and Bourne, P.E. (2009) A unified statistical model to support local sequence order independent similarity searching for ligand-binding sites and its application to genome-based drug discovery. Bioinformatics, 25, i305–i312.]. These algorithms have been extensively benchmarked and shown to outperform most existing algorithms. Moreover, several predictions resulting from SMAP-WS have been validated experimentally. Thus far SMAP-WS has been applied to predict drug side effects, and to repurpose existing drugs for new indications. SMAP-WS provides both a user-friendly web interface and programming API for scientists to address a wide range of compute intense questions in biology and drug discovery.
AVAILABILITY
SMAP-WS is available from the URL http://smap.nbcr.net.
INTRODUCTION
The 3D structure of a protein is an essential component in elucidating biological function(s) at the molecular level. Ligand-binding sites and their interactions with binding partners provides a strong correlation between structure and function and thus are critical to address a wide range of fundamental and practical problems—predicting functions for structural genomics targets, bridging knowledge derived from small molecules and proteins, correlating molecular functions to physiological processes, studying protein evolution and diversity, and designing safe and efficient therapeutics.
The SMAP web service (SMAP-WS) is distinct from the downloadable software and is designed for web accessible 3D ligand-binding site comparison and similarity searching on a structural proteome scale. The underlying algorithms comprising SMAP-WS and the standalone software, SMAP, are distinct from existing web servers SiteEngine (1), SitesBase (2,3), CavBase (4–6), SuMo (7), PdbSiteScan (8), eF-Site (9,10), pvSOAR (11), ProFunc (12), PevoSoar (13) and fPOP(14). First, SMAP represents protein structures using C-α atoms only and hence is tolerant to structural variation, meaning it can be applied to homology models and low-resolution structures. Second, amino acid residues are characterized by surface orientation and a geometric potential (15) which provides a geometrical constraint to reduce the search space when undertaking ligand-binding site comparison. Third, two structures are compared using a sequence order independent profile–profile alignment (SOIPPA) algorithm (16). SOIPPA aligns two structures in the spirit of local sequence alignment, but independent of the sequence order. As a result the location and boundary of the ligand-binding site does not need to be pre-defined. This property is important for real world applications since information on the ligand-binding site may be unknown. Fourth, SMAP can compare two biological units that may include multiple chains. This is important since binding sites may be located in the homo- or hetero-dimer interface. For example, the binding site of the antibiotic myxopyronin to the bacterial RNA polymerase is located in the ‘switch region’ between the β and β′ chains. Finally, SMAP determines the similarity between two binding sites through the combination of geometrical fit, residue conservation and physiochemical similarity. The statistical significance of the similarity is estimated using an extreme value distribution model (17). Putting these features together within a parallel computing environment means that SMAP-WS is capable of an all-by-all comparison of binding sites for a complete structural proteome.
In benchmark studies, SOIPPA outperforms most existing ligand-binding site comparison algorithms (16). Around 30% of evolutionary and functional relationships across superfamilies are identified by SOIPPA with a false–positive ratio of 5%. Moreover, SOIPPA outperforms global structural alignment algorithms in detecting remote homologous that belong to the same superfamily. For a false–positive ratio of 5%, SOIPPA detects 15% more true positives than the global structural alignment. More important, several predictions from SMAP have been experimentally validated (16,18–20). Given the reliability of SMAP, it has been applied to constructing drug–target interaction networks on a structural proteome scale (17), predicting molecular mechanisms of drug side effects (21,22), repurposing old drugs for new medical usage (19), designing polypharmacology (dirty) drugs (18), and establishing evolutionary relationships across protein fold space (16). Thus, SMAP is useful for studying fundamental questions in protein structure, function and evolution, as well as for computer aided drug design based on polypharmacology. As standalone software, SMAP can be installed locally and executed from the command line. SMAP-WS has several improvements that make it more user-friendly and computationally efficient. SMAP-WS has a web-based interface for the input of PDB structures, the set of required parameters, a Jmol visualization plugin to analyze results, pre-computed databases to search against and a parallel implementation of SMAP accessible from a large compute cluster to improve database search speed. Thus SMAP-WS facilitates the application of comparative ligand-binding site analysis to address practical problems in biology and drug discovery.
METHODS
Opal powered SMAP web services
SMAP-WS is powered by Opal (23), a toolkit that enables scientists to easily wrap applications as web services that have user-friendly web forms by configuring simple XML files. Two SMAP-WS interfaces are implemented: (i) pair-wise comparison of two potential ligand-binding sites; and (ii) search using a query structure against a non-redundant structure database from the RCSB Protein Data Bank (PDB) (24). In the first application structures and their components can be chosen from the PDB or uploaded by the user. In the second application the user may either choose to enter a PDB structure id and its chain id(s) or upload a structure file in PDB format. The user can then choose to perform a search using this structure against several databases, including human homologous proteins and non-redundant PDB structures based on sequence identities of 30 and 90%, respectively. The user has the option to modify the appropriate SMAP-WS parameters for both applications. In order to improve the search speed, the database search has pre-cached the protein structures used for the SMAP comparison. The structure cached is characterized by geometric, evolutionary and physiochemical properties and uses default parameters. The pair-wise comparison interface provides the user with the ability to modify more parameters for comparing two protein structures based on the similarity of their potential ligand-binding sites.
In additional to the web input forms (http://smap.ncbr.net), SMAP-WS can be accessed through a programming API. The details of how to write a client program can be found on the web site.
Output of SMAP-WS
The hits from a database search are sorted by the similarity score of the match, along with P-values of the match, their PDB structure ids, chain ids and biological descriptions. The PDB id is linked to the structure summary page of the RCSB PDB (http://www.rcsb.org/pdb). For each of the hits, detailed information on the ligand-binding site similarity is presented (P-value, raw alignment score, RMSD and Tanimoto coefficient of overlap). The amino-acid residue alignment between two ligand-binding sites and the transformation matrix to superpose them are also displayed.
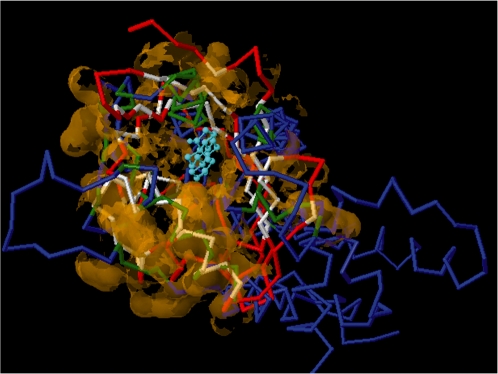
It is important to evaluate if the predicted residue cluster is a potential binding region. SMAP-WS relies on the geometric potential (15), which is a shape descriptor to characterize both local and global topological properties of each residue, to determine whether a residue is located in a pocket on the protein structure or not. However, in a real application where the binding region is unknown, additional information such as ligand-binding affinity may be required to determine if the predicted region is suited for ligand binding. Thus, a visualization tool that allows the user to inspect the protein–ligand complex structure was implemented. A Jmol plugin (Jmol: an open-source Java viewer for chemical structures in 3D.) that displays the superposition of two protein structures with predicted and aligned ligand-binding site residues is provided. An example of such a superposition is shown in Figure 1. The estrogen receptor ligand-binding domain (PDB id: 1QKT) is compared with the steroid delta-isomerase (PDB id: 1OHP) without specifying the co-crystallized ligand-binding site (in the web interface, the option of ‘search only co-crystal ligand sites’ for both structures was set to false). A statistically significant similarity between the two estradiol ligand-binding sites is detected (P = 4.09e–6), although the two structures do not share global structure or sequence similarity [FATCAT (25) P-value 1.19e–1, and sequence identity %9.62], and the estradiol-binding sites are not pre-defined in both of the structures.
Figure 1.
Two superposed structures of steroid delta-isomerase (PDB id: 1OHP, blue backbone) and estrogen receptor ligand-binding domain (PDB id 1QKT, red backbone) from an SMAP alignment. The co-crystalized estradiol in 1QKT is shown as a light blue stick model. The aligned residues between the two structures are highlighted.
One of the major applications of SMAP is to predict off-targets given a known protein–ligand complex. The ligand-binding site similarity between two proteins alone is prerequisite, but not sufficient to determine their cross-reactivity for a specific ligand. The chemical nature of the ligand also contributes to the binding promiscuity. For example, staurosporine can bind to a large panel of kinases with Kd < 100 nm. However, compound VX-745 is a highly specific ATP competitive inhibitor of p38 MAP kinase (26). A more recent case is the chemical phylogenetic study of histone deacetylases (HDAC), where two chemical analogs have different binding profiles across multiple members of the HDAC superfamily (27). To computationally determine the potential off-target binding, it is necessary to calculate the binding free energy of the protein–ligand complex using techniques such as protein–ligand docking and molecular dynamics simulation. SMAP-WS narrows down the potential off-targets to a small subset of the whole structure proteome as well as provides an initial binding pose for the given ligand to the off-target it found in the query structure. The accuracy of predicted binding poses by SMAP-WS has been evaluated in a previous study (16). In a rigorous benchmark test, 6.5 and 25.9% of predicted binding poses fall within RMSD values of <2.0 and <5.0 Å, respectively, when compared with co-crystallized ligands that bind to proteins with different folds. Hence, the predicted protein–ligand complex from SMAP-WS could be used as a starting point for more computational intensive studies. The pipeline has been successfully applied to determine the polypharmacological targets of Trypanosoma Brucei RNA-ligase inhibitors (18). To facilitate such applications, SMAP-WS allows users to download the structure of potential off-targets with the superposed ligand. These complexes can then be subject to more computationally intensive studies such as protein–ligand docking and MD simulation.
Paralleled implementation of SMAP-WS
SMAP-WS database search is scheduled by the Sun Grid Engine (SGE), which allows SMAP pair-wise comparison to be executed concurrently on all available compute nodes. As a result, SMAP-WS significantly improves the speed of ligand-binding site database searching. Using SMAP on a single processor, sequential comparison of a query structure against a database of about 40 000 non-redundant structures from the PDB takes more than 20 days (17). Our solution to speed up this process was to set up a wrapper program that submits a SGE array job for the set of SMAP comparisons to allow these comparisons to run in parallel on the computer nodes in a cluster. The SMAP-WS server cluster has available up to 99 computer nodes with two processors on each node. Thus 198 SMAP-WS jobs can run in parallel, when all computer nodes are available, with a scan of the non-redundant PDB being done within one day.
CONCLUSION
We have developed a high performance computation environment SMAP-WS for protein ligand-binding site comparison and database searching. SMAP-WS provides both user-friendly interfaces and a programming API to help a wide spectrum of scientists to access the service. It is expected that the integration of SMAP-WS with other bioinformatics, molecular modeling and systems biology tools will facilitate the study of protein–ligand interactions on a structural proteome scale and drug design based on polypharmacology.
FUNDING
National Institutes of Health grant GM078596; and National Center for Research Resources, National Institutes of Health NIH P41RR08605 for the National Biomedical Computation Resource (NBCR). Funding for open access charge: National Institutes of Health.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors would like to thank Dr Li Xie for helpful discussions. The authors appreciate the constructive suggestions of the anonymous reviewers and the editor in improving the manuscript.
REFERENCES
- 1.Shulman-Peleg A, Nussinov R, Wolfson HJ. Recognition of functional sites in protein structures. J. Mol. Biol. 2004;339:607–633. doi: 10.1016/j.jmb.2004.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gold ND, Jackson RM. SitesBase: a database for structure-based protein-ligand binding site comparisons. Nucleic Acids Res. 2006;34:D231–D234. doi: 10.1093/nar/gkj062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gold ND, Jackson RM. A searchable database for comparing protein-ligand binding sites for the analysis of structure-function relationships. J. Chem. Inf. Modeling. 2006;46:736–742. doi: 10.1021/ci050359c. [DOI] [PubMed] [Google Scholar]
- 4.Kuhn D, Weskamp N, Schmitt S, Hullermeier E, Klebe G. From the similarity analysis of protein cavities to the functional classification of protein families using cavbase. J. Mol. Biol. 2006;359:1023–1044. doi: 10.1016/j.jmb.2006.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schmitt S, Kuhn D, Klebe G. A new method to detect related function among proteins independent of sequence and fold homology. J. Mol. Biol. 2002;323:387–406. doi: 10.1016/s0022-2836(02)00811-2. [DOI] [PubMed] [Google Scholar]
- 6.Weskamp N, Kuhn D, Hullermeier E, Klebe G. Efficient similarity search in protein structure databases by k-clique hashing. Bioinformatics. 2004;20:1522–1526. doi: 10.1093/bioinformatics/bth113. [DOI] [PubMed] [Google Scholar]
- 7.Jambon M, Imberty A, Deleage G, Geourjon C. A new bioinformatic approach to detect common 3D sites in protein structures. Proteins. 2003;52:137–145. doi: 10.1002/prot.10339. [DOI] [PubMed] [Google Scholar]
- 8.Ivanisenko VA, Pintus SS, Grigorovich DA, Kolchanov NA. PDBSiteScan: a program for searching for active, binding and posttranslational modification sites in the 3D structures of proteins. Nucleic Acids Res. 2004;32:W549–W554. doi: 10.1093/nar/gkh439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kinoshita K, Furui J, Nakamura H. Identification of protein functions from a molecular surface database, eF-site. J. Struct. Func. Genomics. 2002;2:9–22. doi: 10.1023/a:1011318527094. [DOI] [PubMed] [Google Scholar]
- 10.Kinoshita K, Nakamura H. Identification of protein biochemical functions by similarity search using the molecular surface database eF-site. Protein Sci. Pub. Protein Soci. 2003;12:1589–1595. doi: 10.1110/ps.0368703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Binkowski TA, Freeman P, Liang J. pvSOAR: detecting similar surface patterns of pocket and void surfaces of amino acid residues on proteins. Nucleic Acids Res. 2004;32:W555–W558. doi: 10.1093/nar/gkh390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Laskowski RA, Watson JD, Thornton JM. ProFunc: a server for predicting protein function from 3D structure. Nucleic Acids Res. 2005;33:W89–W93. doi: 10.1093/nar/gki414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tseng YY, Dundas J, Liang J. Predicting protein function and binding profile via matching of local evolutionary and geometric surface patterns. J. Mol. Biol. 2009;387:451–464. doi: 10.1016/j.jmb.2008.12.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tseng YY, Chen ZJ, Li WH. fPOP: footprinting functional pockets of proteins by comparative spatial patterns. Nucleic Acids Res. 38:D288–D295. doi: 10.1093/nar/gkp900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xie L, Bourne PE. A robust and efficient algorithm for the shape description of protein structures and its application in predicting ligand binding sites. BMC Bioinformatics. 2007;8(Suppl. 4):S9. doi: 10.1186/1471-2105-8-S4-S9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xie L, Bourne PE. Detecting evolutionary relationships across existing fold space, using sequence order-independent profile-profile alignments. Proc. Natl Acad. Sci. USA. 2008;105:5441–5446. doi: 10.1073/pnas.0704422105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xie L, Xie L, Bourne PE. A unified statistical model to support local sequence order independent similarity searching for ligand-binding sites and its application to genome-based drug discovery. Bioinformatics. 2009;25:i305–i312. doi: 10.1093/bioinformatics/btp220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Durrant JD, Amaro RE, Xie L, Urbaniak MD, Ferguson MA, Haapalainen A, Chen Z, Di Guilmi AM, Wunder F, Bourne PE, et al. A multidimensional strategy to detect polypharmacological targets in the absence of structural and sequence homology. PLoS Comput. Biol. 2010;6:e1000648. doi: 10.1371/journal.pcbi.1000648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kinnings SL, Liu N, Buchmeier N, Tonge PJ, Xie L, Bourne PE. Drug discovery using chemical systems biology: repositioning the safe medicine Comtan to treat multi-drug and extensively drug resistant tuberculosis. PLoS Comput. Biol. 2009;5:e1000423. doi: 10.1371/journal.pcbi.1000423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miller JR, Dunham S, Mochalkin I, Banotai C, Bowman M, Buist S, Dunkle B, Hanna D, Harwood HJ, Huband MD, et al. A class of selective antibacterials derived from a protein kinase inhibitor pharmacophore. Proc. Natl Acad. Sci. USA. 2009;106:1737–1742. doi: 10.1073/pnas.0811275106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xie L, Li J, Xie L, Bourne PE. Drug discovery using chemical systems biology: identification of the protein-ligand binding network to explain the side effects of CETP inhibitors. PLoS Comput. Biol. 2009;5:e1000387. doi: 10.1371/journal.pcbi.1000387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xie L, Wang J, Bourne PE. In silico elucidation of the molecular mechanism defining the adverse effect of selective estrogen receptor modulators. PLoS Comput. Biol. 2007;3:e217. doi: 10.1371/journal.pcbi.0030217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Krishnan S, Clementi L, Ren J, Papadopoulos P, Li W. The 2009 IEEE Congress on Services (SERVICES-1 2009), July, 2009. Los Angeles, CA, USA: IEEE Conference; 2006. Design and Evaluation of Opal2: A Toolkit for Scientific Software as a Service. [Google Scholar]
- 24.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ye Y, Godzik A. FATCAT: a web server for flexible structure comparison and structure similarity searching. Nucleic Acids Res. 2004;32:W582–W585. doi: 10.1093/nar/gkh430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Karaman MW, Herrgard S, Treiber DK, Gallant P, Atteridge CE, Campbell BT, Chan KW, Ciceri P, Davis MI, Edeen PT, et al. A quantitative analysis of kinase inhibitor selectivity. Nature Biotechnol. 2008;26:127–132. doi: 10.1038/nbt1358. [DOI] [PubMed] [Google Scholar]
- 27.Bradner JE, West N, Grachan ML, Greenberg EF, Haggarty SJ, Warnow T, Mazitschek R. Chemical phylogenetics of histone deacetylases. Nature Chem. Biol. 2010;6:238–243. doi: 10.1038/nchembio.313. [DOI] [PMC free article] [PubMed] [Google Scholar]