Abstract
The productivity of any scientist is affected by cumbersome, tedious and time-consuming tasks that try to make the heterogeneous web services compatible so that they can be useful in their research. MOWServ, the bioinformatic platform offered by the Spanish National Institute of Bioinformatics, was released to provide integrated access to databases and analytical tools. Since its release, the number of available services has grown dramatically, and it has become one of the main contributors of registered services in the EMBRACE Biocatalogue. The ontology that enables most of the web-service compatibility has been curated, improved and extended. The service discovery has been greatly enhanced by Magallanes software and biodataSF. User data are securely stored on the main server by an authentication protocol that enables the monitoring of current or already-finished user’s tasks, as well as the pipelining of successive data processing services. The BioMoby standard has been greatly extended with the new features included in the MOWServ, such as management of additional information (metadata such as extended descriptions, keywords and datafile examples), a qualified registry, error handling, asynchronous services and service replication. All of them have increased the MOWServ service quality, usability and robustness. MOWServ is available at http://www.inab.org/MOWServ/ and has a mirror at http://www.bitlab-es.com/MOWServ/.
INTRODUCTION
Diversity, heterogeneity and geographical dispersion of biological data constitute problems that hinder the potential integration of such information. Therefore, researcher’s productivity is affected by tedious, time-consuming and prone-to-error tasks such as searching for the appropriate web services, collecting URLs, familiarizing themselves with the different service interfaces, transferring data from one service to another, formatting data for compatibility purposes or copy/paste data in web-forms with different interfaces, to mention a few. The development of systems for interprocess communication has been previously carried out with different goals: gathering multiple services with reliable access (1), providing access to a collection of independent analysis tools (2,3) or enabling the communication between a reduced set of tools (4–7). Standardization of bioinformatics services has also been largely analysed (8–15), standing-up over them the use of web-services designed to support automatic machine-to-machine interaction over a network, representing BioMoby (16) the more successful case. In fact, the development of low-level data-interchange methods based on a specific ontology, together with the ability for wiring services to build powerful bioinformatic machines, has been revealed as the most promising solution (17) as the growing number of web-based services for integrating bioinformatic tools demonstrates. MOWServ (18), the bioinformatic platform offered by the Spanish National Institute of Bioinformatics (INB), provides an integrated access to databases and analytical tools and has strongly contributed to the development of the standard BioMoby protocol (17). In this article, the extended capabilities of MOWServ are presented. The new functionality is related to web-services discovering, automatic support for data handling and standardization, and additional protocols to extend the BioMoby scope, and the MOWServ service quality, usability and robustness.
NEW CHARACTERISTICS OF MOWServ
Since the public release of MOWServ (18), the number of available services stands at around 600, and MOWServ invocations have also grown, managing more than 1 million in the last year. Since EMBRACE (19) is part of the Biocatalogue (http://www.biocatalogue.org) that has become the repository of reference for web-services in life Sciences, MOWServ web-services have also been registered there, representing about 50% of the total web services in the registry.
The increase of MOWServ capabilities has been possible due to the following improvements.
New web-service registry
Web-service registration policy at MOWServ is more restrictive than for other BioMoby registries: new services should be registered at the development server (moby-dev, http://moby-dev.inab.org/MOWServ), where additional functional and technical information, as well as examples of input and output objects, has to be included. A form containing all mandatory information for MOWServ (but not in BioMoby standard) is required to be filled in. Only service owners can modify or remove their registration through the web interface or the BioMoby agent provided when they were registered at MOWServ. Service registry obliges service providers to test their service on the development server before final validation regarding ontology localization and correct functioning in line with INB standards (e.g. information to generate help pages and the use of true XML objects as input and output data types). This procedure aims to guarantee the reliability of INB services. Validated services are then moved to the main server (http://www.inab.org/MOWServ) and its mirrors (like http://www.bitlab-es.com/MOWServ/) and are immediately made available to the world. As a matter of fact, users can make use of web services in the moby-dev server instead of the central one. However this is not recommended since moby-dev located services may not be completely functional or comply with INB rules. An additional way of guaranteeing service quality is the daily testing of service availability by the MOWServ. These checks can also be requested and performed on demand through the user interface.
Ontology modifications
The initial version of MOWServ (18) used the Canadian MOBY Central ontology, but the need for standardization soon became evident. The controlled protocol for registration of new services and data types implemented in MOWServ revealed a more coherent organization of resources while guaranteeing compatibility with MOBY Central. It classifies web services into two main groups: basic services that simply manipulate objects (e.g. to create, to convert, to parse or to display them) are in the ‘Object Handling’ branch, and true bioinformatics algorithms are grouped in the ‘Bioinformatics’ branch of the ontology. The service name is intended to be self-explanatory and follows intuitive nomenclature rules (runClustalwFast, getNucleotideSequence, getBestHitsFromBlast). Likewise, additional metadata requested during the registration procedure allows MOWServ to provide detailed informative support in the form of tooltips or in the service interface (at parameter level). Apart from the well-known Blast, Fasta, ClustalW, Phylip, T-Coffee and EMBOSS applications, the Bioinformatics branch includes the specific offer of web services developed by the different research nodes of the INB. Regarding the Object Handling branch, it includes a new node named ‘Displaying’ that gathers web services for image visualization. Datatypes ontology was also simplified while maintaining compatibility with Canada MOBY Central (17).
Scheduler
Due to geographic dispersion, possible multiple instances of the same service and the large number of clients that have access to the MOWServ, a mechanism to improve the reliability of service invocations (the execution of tasks), became necessary. The scheduler module in the MOWServ can be plugged with specific strategies in order to distribute the computational load among the different mirrors. Currently, the default option is to select the mirror in a round-robin fashion. The scheduler module in the MOWServ thereby enhances the robustness and fault tolerance of service invocations.
Asynchronism
BioMoby uses HTTP as a transport protocol; however, this can lead to time-outs and breaks in client–server communication for long-running service invocations. The working solution has been to split the service invocation into several communications between client and service (asynchronous communication). Therefore, results must be temporarily stored locally by services until clients can retrieve them. This asynchronism protocol, which is now part of the BioMoby standard as a SOAP feature (17), is available to all users. Many of the available INB web services are able to communicate using this protocol, and MOWServ fully supports communication with asynchronous services.
Error handling
The initial BioMoby API approach to handling errors consisted of returning an empty package without any other information. Users and developers had no information when a service execution failed. The inclusion of a scheduler (see above) underscores this limitation since the source of error (mainly failed network communication and server down events) must be known in order to choose another mirror to execute pending tasks. Errors can be critical (no result produced) or a warning (an incomplete result can be produced), and can be accompanied by useful information for users and developers. Whatever the error was, a human-readable text with a little description of the problem is returned along with a numerical code to identify and handle errors automatically.
Advanced search
A list of compatible web services that accept a selected object stored in the user’s account has been available from the beginning of the MOWServ (18). This helps users discover new web services based on the data they wish to process and also facilitates integration of different services. As mentioned above, efforts were made to create well-designed ontology to simplify service discovery. However, service discovery has become more difficult since the MOWServ platform has grown to include hundreds of bioinformatics web services and many specialized data types.
For this reason, great effort has been made to facilitate service discovery. Users can search for a word in the service and data type ontology branches, which mark the matches directly in the tree. For more complex service discovery, the MOWServ has been extended with a new discovery tab based on the Magallanes system (20). Magallanes is a search engine enabled to use metadata from various service metadata repositories, including the INB service catalogues. Users specify a query (for example, a word), which is matched against service and data type descriptions. These matches can be exact or approximated using Levenshtein’s text distance. If a search term does not exactly match any service, the discovery module automatically suggests other search terms (as ‘did you mean?’ suggestions). Results are ranked not only by their degree of matching with the search term, but also based on user selections.
In the same way that the number of services has been increased, the number of files hosted in the MOWServ per user has also risen in the last years. To help users find the right object, the file explorer has been remodelled. Using Flex technology, users can now go through the complete list of objects and order the elements by object id, task id, BioMoby type, File name or Date. These fields are also used to filter the information by date.
Pipeline management
Many typical bioinformatics tasks involve the execution of several services in a pipeline. This can be done with the MOWServ by executing one service and then selecting another service to further process the output. However, such pipelines can be automated with the MOWServ. Using the MyGrid’s client Taverna (21) users can create specifications on how individual services should be combined (i.e. the specification of the pipeline). These pipelines can be uploaded to the MOWServ in the users’ accounts and later enacted (executed). Note that the MOWServ limits the pipelines allowed according to a set of requirements (Taverna 1.4 compatibility and only BioMoby services). Pipelines will benefit from all the computational power available in the consortium: two very high-throughput multiprocessors, and the diverse set of services. Earlier versions of MOWServ executed pipelines in a ‘black-box’ mode using Taverna. Currently, parsing of Taverna pipelines is still handled by the Taverna code, but enactment is handled by MOWServ. This means that the pipelines are made more robust in the sense that they can use the replicated BioMoby services in INB (several servers provide independent and redundant services). The MOWServ enactor also requires fewer resources than the previous implementation since the pipelines are split into lightweight processes during execution.
Improved data editing
The process of creating and editing user data has also been improved by a new graphical interface allowing users to create new data with the object editor. The editor now simplifies the editing of some particular types of data such as sequences. Traditionally, BioMoby sequences contain the sequence data itself and the length. The updated editor automatically calculates the length when creating the BioMoby formatted data. Any data created using this editor can be submitted to compatible services later on.
Large sets of biological data (data sets with similar data) can also be edited with a drag-and-drop interface. Users select the existing data objects they have in order to create so-called collections, which can then be sent to compatible services. The collection editor can be used to create sub-collections out of existing collections.
RESULTS
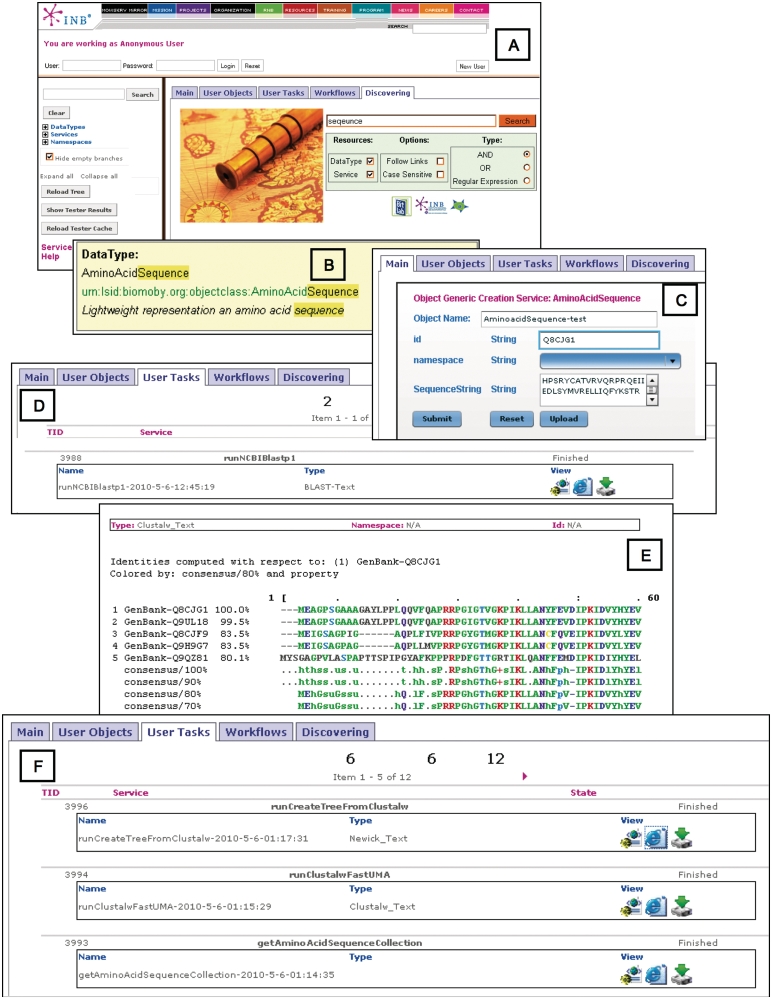
To better demonstrate the MOWServ capabilities, we will walk through a typical user session (Figure 1). Detailed information on this exercise is available as Supplementary Data (http://www.bitlab-es.com/gnv5), and comprehensive material for training is available at the MOWServ web page. In sequence analysis, researchers are frequently faced with the problem of having to discover whether their study protein has been isolated in any new sequenced species or whether new variants have appeared in the protein databanks. Let us assume that we start with a protein sequence on the clipboard.
Figure 1.
Pipeline main steps for a phylogenetic study using web services in MOWServ. (A) Search in Magallanes for a keyword in order to locate the appropriate data type. (B) Detection of a desirable data type, highlighting the keyword matches. (C) Web interface to compose the object corresponding to the selected data type. (D) Monitoring executions by means of the ‘User tasks’ tab, where service Blastp is shown as ‘Finished’. From this tab, the partial results can be visualized in different formats. (E) Specific viewer for an intermediate result in the pipeline (ClustalW). (F) Execution list showing service status.
To proceed in the standard way, identification of the data type related to a keyword such as ‘sequence’ (or similar description of user data) is required. The Magallanes engine is able to perform this task (Figure 1A). Even the ‘Did you mean?’ module in Magallanes could manage spelling mistakes (e.g. ‘seqeunce’) and suggest a number of alternatives. The AminoAcidSequence data type will be among the results (Figure 1B). A simple click over the data type will activate the data editor GUI which, in turn, allows the user to copy and paste from the clipboard and save as an XML file (Figure 1C). A default name is suggested by MOWServ for the new object that can be modified by the user.
The new object will appear in the MOWServ user objects tab and from here the user can request the compatible services for the file in question (clicking on the ‘get compatible services’ button). The runNCBIBlastp1 service, which provides a blast report, appears on the list and the service parameters interface appears when selected, allowing users to execute the service directly using the previously created objects. The service parameter interface is endowed with traditional user-friendly capabilities, such as a list of the more recently used objects, default values for parameters, on-the-fly object creation, automatic naming of output objects, etc. Execution progress can be traced using the ‘user task’ tab (Figure 1D and F), and the new object will be available in the ‘user objects’ tab after reloading the page. The output from runNCBIBlastp1 can be viewed (Figure 1D and E) and further analysed using pipelining capabilities to call getBestHitsFromBlast service in order to extract identifiers from the best hits of the blast report.
It is noteworthy to observe that an alternative process could be started by retrieving a sequence from its ID, using, for instance, the seqID to return the ‘Sequence’. This model can be named single-input/single-output service. When this kind of service is invoked using a collection (multiple-input) of sequence IDs, MOWServ is able to split the collection into individual items, call the services and compose the results again. In this way, the output from getBestHitsFromBlast (a collection), can be used to execute getAminoAcidsSequenceCollection service that invokes getAminoAcidSequence service as many times as sequences are in the collection. The result will be a collection of AminoAcidSequences. Similarity among the sequences gathered in the object can be calculated with the runClustalwFastUMA service that produces a coloured sequence alignment that could be sent to the runCreateTreeFromClustal service in order to group the sequences on the basis of their phylogenetic relationship.
DISCUSSION
BioMoby (17,13) is emerging as the standard of fact for data exchange and web services inter-communication in bioinformatics. MOWServ is a BioMoby-based web client that enables the secure and integrated analysis of data and straightforward access to databases, services and computational resources. Originally endowed with authentication mechanisms, task tracking, automatic and uniform generation of services interfaces, it has been extended with a new formal registry procedure, improved ontology and data edition capabilities, new search tools, error handling, scheduling and asynchronism service invocation. Therefore, traditionally limited CPU-power available at wet-labs has been enhanced by linking together a set of scattered instruments into a single virtual infrastructure, in order to foster more effective and productive research.
The registry procedure aims to guarantee the quality and availability of the services. The controlled registration reduces the risk of broken links, becomes fault-tolerant and offers a homogeneous view of the services and documentation on offer, independently of the status of particular servers. The new ontology offers a more intuitive organization of data types and web services with more nodes and less redundancy, since duplicated objects that populate ancient BioMoby ontologies no longer exist. The new advanced search tool based on Magallanes (20) has been integrated into the MOWServ to facilitate the finding data types and services in the ontology using an efficient algorithm based on inexact matches. The BioMoby API has been extended with asynchronism, error handling and mirroring capabilities both to cope with long-CPU services and to drive executions among the different servers that contain mirrored services. Using asynchronism, clients can retrieve status or results of individual jobs without waiting for the entire service invocation to finish thus avoiding frequent time-outs in web connections. Much of the functionality of the MOWServ is also available in a desktop-based application developed by our group called jOrca (22).
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
The National Institute for Bioinformatics, a platform of the Genoma-España Foundation; the European Union co-funded project ‘Advancing Clinico-Genomic Trials on Cancer’ (contract-026996); Red de Investigación de Reacciones Adversas a Alergenos y Fármacos – Redes Tematicas de Investigación Cooperativa Sanitaria (RIFAAF – RETICS RD07/0064/0017); Ministerio de Ciencia e Innovación, proyecto I+D AGL2009-12139-C02-02; Universidad Pablo de Olavide (CABD-CSIC/JA/UPO).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors would like to thank all the other nodes in the INB infrastructure for their close involvement in the development of the services and integration procedures.
REFERENCES
- 1.Gasteiger E, Gattiker A, Hoogland C, Ivanyi I, Appel RD, Bairoch A. Expasy: the proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 2003;31:3784–3788. doi: 10.1093/nar/gkg563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Basu MK. Sewer: a customizable and integrated dynamic html interface to bioinformatics services. Bioinformatics. 2001;17:577–578. doi: 10.1093/bioinformatics/17.6.577. [DOI] [PubMed] [Google Scholar]
- 3.Rost B, Yachdav G, Liu J. The predictprotein server. Nucleic Acids Res. 2004;32:W321–W326. doi: 10.1093/nar/gkh377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Douguet D, Labesse G. Easier threading through web-based comparisons and cross-validations. Bioinformatics. 2001;17:752–753. doi: 10.1093/bioinformatics/17.8.752. [DOI] [PubMed] [Google Scholar]
- 5.Gracy J, Chiche L. Pat: a protein analysis toolkit for integrated biocomputing on the web. Nucleic Acids Res. 2005;33:W65–W71. doi: 10.1093/nar/gki455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Letondal C. A web interface generator for molecular biology programs in unix. Bioinformatics. 2001;17:73–82. doi: 10.1093/bioinformatics/17.1.73. [DOI] [PubMed] [Google Scholar]
- 7.Perriere G, Combet C, Penel S, Blanchet C, Thioulouse J, Geourjon C, Grassot J, Charavay C, Gouy M, Duret L, et al. Integrated databanks access and sequence/structure analysis services at the pbil. Nucleic Acids Res. 2003;31:3393–3399. doi: 10.1093/nar/gkg530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Aldana JF, Hidalgo-Conde M, Navas I, Roldán MM, Trelles O. IADIS International Conference, Applied Computing. 2005. Bio-broker: a biological data and services mediator system. 2005, Algarve, Portugal. [Google Scholar]
- 9.Badidi E, De Sousa C, Lang BF, Burger G. Anabench: a web/corba-based workbench for biomolecular sequence analysis. BMC Bioinform. 2003;4:63. doi: 10.1186/1471-2105-4-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carrere S, Gouzy J. Remora: a pilot in the ocean of biomoby web-services. Bioinformatics. 2006;22:900–901. doi: 10.1093/bioinformatics/btl001. [DOI] [PubMed] [Google Scholar]
- 11.de Knikker R, Guo Y, Li JL, Kwan AK, Yip KY, Cheung DW, Cheung KH. A web services choreography scenario for interoperating bioinformatics applications. BMC Bioinform. 2004;5:25. doi: 10.1186/1471-2105-5-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stevens RD, Robinson AJ, Goble CA. mygrid: personalised bioinformatics on the information grid. Bioinform. 2003;19(Suppl. 1):i302–i304. doi: 10.1093/bioinformatics/btg1041. [DOI] [PubMed] [Google Scholar]
- 13.Wilkinson M, Schoof H, Ernst R, Haase D. Biomoby successfully integrates distributed heterogeneous bioinformatics web services. the planet exemplar case. Plant Physiol. 2005;138:5–17. doi: 10.1104/pp.104.059170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Soh J, Gordon PMK, Taschuk ML, Dong A, Ah-Seng AC, Turinsky AL, Sensen CL. Bluejay 1.0: genome browsing and comparison with rich customization provision and dynamic resource linking. BMC Bioinform. 2008;9:450. doi: 10.1186/1471-2105-9-450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen Y, Lawless C, Gillespie CS, Wu J, Boys RJ, Wilkinson DJ. Calibayes and basis: integrated tools for the calibration, simulation and storage of biological simulation models. Brief Bioinform. 2010;11:278–289. doi: 10.1093/bib/bbp072. [DOI] [PubMed] [Google Scholar]
- 16.Wilkinson MD, Links M. Biomoby: an open source biological web services proposal. Brief Bioinform. 2002;3:331–341. doi: 10.1093/bib/3.4.331. [DOI] [PubMed] [Google Scholar]
- 17.Wilkinson MD, Senger M, Kawas E, Bruskiewich R, Gouzy J, Noirot C, Bardou P, Ng A, Haase D, de Andres Saiz E, et al. Interoperability with moby 1.0–it’s better than sharing your toothbrush! Brief Bioinform. 2008;9:220–231. doi: 10.1093/bib/bbn003. [DOI] [PubMed] [Google Scholar]
- 18.Navas-Delgado I, Rojano-Muñoz MDM, Ramìrez S, Pérez AJ, León EA, Aldana-Montes JF, Trelles O. Intelligent client for integrating bioinformatics services. Bioinformatics. 2006;22:106–111. doi: 10.1093/bioinformatics/bti740. [DOI] [PubMed] [Google Scholar]
- 19.Pettifer S, Thorne D, McDermott P, Attwood T, Baran J, Bryne JC, Hupponen T, Mowbray D, Vriend G. An active registry for bioinformatics web services. Bioinformatics. 2003;25:2090–2091. doi: 10.1093/bioinformatics/btp329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rìos J, Karlsson J, Trelles O. Magallanes: a web services discovery and automatic workflow composition tool. BMC Bioinform. 2009;10:334. doi: 10.1186/1471-2105-10-334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Oinn T, Addis M, Ferris J, Marvin D, Senger M, Greenwood M, Carver T, Glover K, Pocock MR, Wipat A, et al. Taverna: a tool for the composition and enactment of bioinformatics workflows. Bioinformatics. 2004;20:3045–3054. doi: 10.1093/bioinformatics/bth361. [DOI] [PubMed] [Google Scholar]
- 22.Martín-Requena V, Ríos J, García M, Ramírez S, Trelles O. jorca: easily integrating bioinformatics web services. Bioinformatics. 2010;26:553–559. doi: 10.1093/bioinformatics/btp709. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.