Abstract
New sequencing technologies, such as Roche 454, ABI SOLiD and Illumina, have been increasingly developed at an astounding pace with the advantages of high throughput, reduced time and cost. To satisfy the impending need for deciphering the large-scale data generated from next-generation sequencing, an integrated software MagicViewer is developed to easily visualize short read mapping, identify and annotate genetic variation based on the reference genome. MagicViewer provides a user-friendly environment in which large-scale short reads can be displayed in a zoomable interface under user-defined color scheme through an operating system-independent manner. Meanwhile, it also holds a versatile computational pipeline for genetic variation detection, filtration, annotation and visualization, providing details of search option, functional classification, subset selection, sequence association and primer design. In conclusion, MagicViewer is a sophisticated assembly visualization and genetic variation annotation tool for next-generation sequencing data, which can be widely used in a variety of sequencing-based researches, including genome re-sequencing and transcriptome studies. MagicViewer is freely available at http://bioinformatics.zj.cn/magicviewer/.
INTRODUCTION
New sequencing technologies, such as Roche 454, ABI SOLiD and Illumina Solexa, have been widely applied to various aspects of biological researches, with advantages of reduced time, cost and efforts (1,2). These technologies have generated unprecedented amounts of sequence reads. A single run of the Illumina Genome Analyzer II, for example, can generate terabytes of data and >10 gigabases of raw reads. Therefore, great efforts should be made to alter the status quo that specialized analysis tools for sequencing data are lagged behind next-generation sequencing technologies, which to a certain extent, directly restricts the efficiency of handling and manipulating such a large amount of data. From this perspective, bioinformatics approaches and software are encountering great challenges of handling and analyzing the large-scale data generated from genome-wide studies (3,4).
Recently, several bioinformatics tools specifically designed for visualizing next-generation sequencing data have been introduced toward facilitating the interpretation of large-scale data. EagleView is developed to visualize large genome assemblies of next-generation sequence reads with an input of ACE format (5). LookSeq, an AJAX-based web tool, can display large data sets of aligned sequence reads and multiple layers of information contained in deep sequencing data (6). MapView is developed to present hundreds of millions of short read alignment and detect genetic variation on a desktop computer with a MVF input format (7). NGSView is an extensible open source sequence editor for visualization and manipulation of a massive amount of short reads (8). Tablet is a dedicated software designed for next-generation sequencing data visualization, with a range of input assembly formats (9). BamView is a Java application designed for visualizing large amounts of short reads alignment with an input of BAM (Binary Alignment/Map) format (10). inGAP, albeit not specifically designed for sequencing data visualization, can map short reads to reference genome, detect and annotate genetic variation and assist genome assembly through a user-friendly graphical interface (11).
In general, an ideal next-generation sequencing data visualization tool is supposed to be fast and memory efficient. Considering the fast growth of next-generation sequencing technologies, speed and memory usage should be the most important factor in building software. A customizable interface comes as a second, in which various aspects of information can be easily accessed by users through an adjustable interface as well as color-definable settings (3). Moreover, it should be platform independent, easy to use and compatible with other software. Such qualified software will make the analytic process as efficient and painless as possible and deserve to be an authentic next-generation integrated solution for data analysis.
Herein, to satisfy the impending need for deciphering large-scale data generated by next-generation sequencing, an integrated software MagicViewer is developed to visualize short read alignment, identify and annotate genetic variation based on the reference genome sequence. As an integrated solution, MagicViewer can serve as a visualization tool to display large-scale reads, which is featured with operating system independence, user-friendly interface, multiple navigation views, zoom mode and customized color schemes. Another feature of MagicViewer is that it provides extensive options for users to detect, filter and annotate genetic variation between short reads and reference genomes. From our experiments, sample applications are provided to demonstrate how MagicViewer facilitates the large-scale data interpretation as convenient and effortless as possible in the face of next-generation sequencing data.
DATA INPUT
Different tools often use their own defined format for the data input (e.g. XML format was used as input for NGSView and MVF format for MapView, etc.), which shows the weakness of the compatibility and leads to laborious efforts to convert various formats. This contradiction is especially prominent when processing huge mass of data obtained from high-throughput sequencing. Most recently, a generic alignment (SAM) format has been developed for storing aligned short reads in a flexible style with compacted size (12). Hence, to be compatible with such a powerful format, MagicViewer employed the SAM format to enable an easy conversion of various input file formats, including PSL, MAQ, Bowtie, SOAP and ZOOM.
When starting with a new project, MagicViewer requires a reference genome sequence in fasta format, a sorted bam file containing the aligned short reads obtained from SAMtools (12) and an optional reference genome annotation file in GFF format. MagicViewer can save intermediate results as a log file, thereby facilitating an easier manipulation of project for later reuse. Taking existing archive into account, MagicViewer introduces a conspicuously new feature of workspace where users can load their most frequently used resources for quick access. Through such a convenient way, users can easily load, browse, further update and modify their previous results, instead of reconstructing a new project.
ALIGNMENT VISUALIZATION
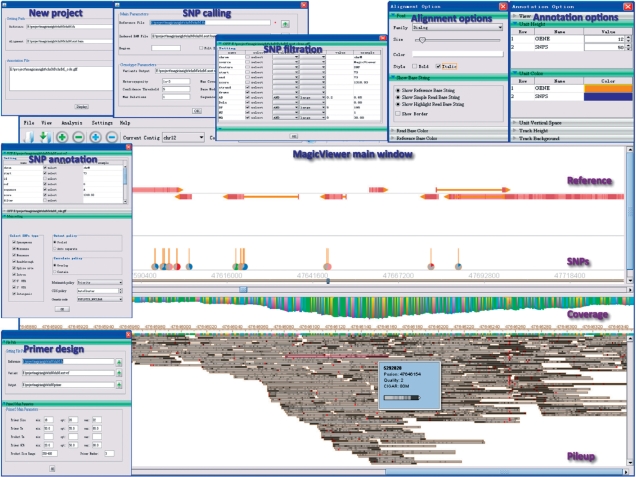
MagicViewer, written in the Java programming language, provides a user-friendly interface and can be performed in a standalone, operating system-independent manner (Figure 1). Large-scale short reads mapped onto the reference genome are optimally placed in multiple lines with compact arrangement and can be visualized intuitively. To get a better graph view, users can acquire scrollable thumbnail image through zooming in and out. Theoretically, the short reads image can be zoomed to any resolution, from whole chromosome to individual bases at any desired level. When the mouse hovers on a specific read, auxiliary information will be shown in a tooltip, such as reads ID, location, base quality, read length and orientation. The sequencing depth distribution of mapped reads can be visualized on the top of graphical representation of short reads alignments. In addition, MagicViewer provides extensive flexibility to change the appearance of the displayed short read alignment and sequencing depth. Users can change font and colors in many different combinations, such as nucleotide and background color. Such a color or format setting function is not trivial, because users usually need a better display when exploring SNPs from hundreds of fold coverage of short read alignments.
Figure 1.
The workflow and screenshots of MagicViewer.
GENETIC VARIATION DETECTION, FILTRATION AND ANNOTATION
Next-generation sequencing technologies have been widely used for effective, easy and in-depth investigation of genetic variation, including SNPs and InDels (insertion/deletions), to a better understanding of human health (13). To satisfy these requirements, beyond a sophisticated short read visualization tool, MagicViewer is devoted to serve as a comprehensive workflow for genetic variation detection, filtration and annotation (Figure 1).
In order to efficiently identify genetic variation between large-scale short reads and reference genomes, the Genome Analysis Toolkit (GATK, http://www.broadinstitute.org/gsa/wiki/index.php/The_Genome_Analysis_Toolkit) is incorporated. The GATK is a structured software library designed to enable rapid development of efficient and robust analysis tools for next-generation sequencing data. The MagicViewer user interface allows users to change many of the parameters, such as heterozygosity, confidence threshold and max coverage. The output of genetic variation calling is organized in a variable call format (VCF, http://www.broadinstitute.org/gsa/wiki/index.php/VCF_Validator), which is the standard variant calling file format used by the 1000 Genomes Project. Meanwhile, the identified genetic variations will be displayed at the top of the main window of MagicViewer for easily interpreting the results. For candidate SNPs, MagicViewer provides a number of versatile display and filtering options for users to remove low confidence SNPs. Such options include thresholds for coverage, quality, variant frequency and number of reads.
Another predominate feature of MagicViewer is that it can be used to link the detected genetic variations to the annotation information of the reference genome. Users need to provide such information in a general feature format (GFF format, http://www.sanger.ac.uk/Software/formats/GFF) before launching a new project. MagicViewer also provides a variety of genetic variation analysis functions, including SNP category selection, result organization and visualization. Users can select a subset of different SNPs categories through custom setting to achieve an extensive annotation, including intergenic, intron, missense, nonsense, readthrough, splice site, synonymous, 3′- and 5′-UTR. The graphical display of output can be customized using supplied options to define view mode, arrow mode, read height, color, vertical space, track display height and background color.
In many genetic variation projects, Sanger sequencing is usually necessary for the verification of the detected genetic variations. Therefore, MagicViewer has also provided a facility to help users design primers for specific genomic region flanking the SNP site in a batch mode by the implement of Primer3 (14). To fulfill this function, MagicViewer allows users to adjust a number of important parameters, including primer length, Tm, GC content, product Tm and the number of primers.
CASE STUDIES
To determine the effectiveness of MagicViewer, we simulated 50 million (75-bp Illumina paired-end reads) with ∼25-fold coverage and 0.001% divergence from human chromosome 8 using the MAQ program (15). As a result, MagicViewer identified a total of 138 604 SNPs with an accuracy of 97.23% in comparison with the original simulated SNPs. By further observation of the undetected SNPs, we found that the majority of them (98.65%) located in repeats or low coverage regions.
In real data sets, MagicViewer was applied to five pooled human exon samples, which were obtained using the NimbleGen 2.1M human exome array and Illumina Genome Analyzer IIx instrument (data not shown). Originally, a total of approximate 56.41 million single-end 75-bp reads were obtained with a size of 3462 MB, among which over 46.12 million reads (35% reads were on targeted exon regions) could be mapped onto the reference genome using the SOAP program with the default setting (data not shown). Among the five pooled samples, MagicViewer identified a number of 28 328 SNPs in targeted exon regions based on default settings, among which homozygous mutations account for 5.53% of the total SNPs and the remaining parts are heterozygous alleles. Functional annotation of these SNPs indicated that synonymous mutations accounted for approximate 18.7%, and the other kinds of mutations were as follows: non-synoymous (31.5%), nonsense (45.2%) and readthrough (4.3%). To experimentally evaluate the robustness of MagicViewer, 80 SNPs were randomly selected for validation using Sequenom’s MassARRAY system (data not shown), and 77 of them were confirmed, revealing the accuracy of MagicViewer in identifying genetic variants.
COMPUTER PERFORMANCE
With an increasing data size generated from high-throughput sequencing, effective resource management is essential for alignment visualization. To enhance the computational efficiency, MagicViewer stores the data in a cache and keeps in memory only the information that is visualized. Here, we tested it on a typical Windows system with Intel Core 2 Duo E7400 (2.80 GHz) and 2 GB memory. We firstly tested it on a small data set (a 175 MB BAM file and a 310 KB reference genome), and found that MagicViewer spent 0.141 s and required approximate 70 MB of RAM to load the data. In addition, extra 2 min 11 s and 16.7 MB of RAM were used to identify 2935 SNPs; 0.746 s and 7.5 MB of RAM for SNP annotation. We also tested it on human genome data (2.57 GB BAM file and 2.95 GB reference genome), which needed approximate 2 s and 100 MB of RAM to load all the input files. Additionally, it took MagicViewer 3 h 30 min 34 s to identify SNPs from the assembly with a 27 MB RAM usage. For the annotation process, MagicViewer required 9 min 51 s and 164 MB of RAM.
PERSPECTIVES
Our main objective in developing MagicViewer is to provide a streamlined framework for short read alignment visualization, genetic variation detection and annotation through a user-friendly graphical display. MagicViewer not only provides users a simple and straightforward way to access large-scale data sets, but also integrates a sophisticated package to annotate genetic variations, which is customizable for researchers to analyze next generation data more efficiently and flexibly. In addition, MagicViewer is implemented in Java environment to ensure portability across multiple platforms. Development of MagicViewer is an ongoing process, and it will be updated and extended when new tools or technologies are available. Currently, MagicViewer only provides detailed annotation facilities for SNPs. Extending annotations for InDels will come out soon. Multiple samples comparison tools are being developed in order to allow MagicViewer to conduct comparison of sequencing depth and genetic variation among various samples. In the future, MagicViewer will provide a query system to allow users to search a particular string of sequences in both reference genome and short reads. MagicViewer is intended to be further improved through user feedbacks. In conclusion, MagicViewer is a powerful short read visualization and genetic variation annotation tool for next-generation sequencing data, which can be widely used in a variety of genome-wide studies, such as de novo sequencing, targeted re-sequencing and transcriptome sequencing.
FUNDING
Funding for open access charge: National High Technology Research and Development Program of China (2006AA02A304); Major State Basic Research Development Program of China (2007CB512302); Natural Science Foundation of Zhejiang Province (Y2090694).
Conflict of interest statement. None declared.
REFERENCES
- 1.Shendure J, Ji H. Next-generation DNA sequencing. Nat. Biotechnol. 2008;26:1135–1145. doi: 10.1038/nbt1486. [DOI] [PubMed] [Google Scholar]
- 2.Ansorge WJ. Next-generation DNA sequencing techniques. Nat. Biotechnol. 2009;25:195–203. doi: 10.1016/j.nbt.2008.12.009. [DOI] [PubMed] [Google Scholar]
- 3.McPherson JD. Next-generation gap. Nat. Methods. 2009;6:S2–S5. doi: 10.1038/nmeth.f.268. [DOI] [PubMed] [Google Scholar]
- 4.Horner DS, Pavesi G, Castrignano T, De Meo PD, Liuni S, Sammeth M, Picardi E, Pesole G. Bioinformatics approaches for genomics and post genomics applications of next-generation sequencing. Brief Bioinform. 2010;11:181–197. doi: 10.1093/bib/bbp046. [DOI] [PubMed] [Google Scholar]
- 5.Huang W, Marth G. EagleView: a genome assembly viewer for next-generation sequencing technologies. Genome Res. 2008;18:1538–1543. doi: 10.1101/gr.076067.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Manske HM, Kwiatkowski DP. LookSeq: a browser-based viewer for deep sequencing data. Genome Res. 2009;19:2125–2132. doi: 10.1101/gr.093443.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bao H, Guo H, Wang J, Zhou R, Lu X, Shi S. MapView: visualization of short reads alignment on a desktop computer. Bioinformatics. 2009;25:1554–1555. doi: 10.1093/bioinformatics/btp255. [DOI] [PubMed] [Google Scholar]
- 8.Arner E, Hayashizaki Y, Daub CO. NGSView: an extensible open source editor for next-generation sequencing data. Bioinformatics. 2010;26:125–126. doi: 10.1093/bioinformatics/btp611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Milne I, Bayer M, Cardle L, Shaw P, Stephen G, Wright F, Marshall D. Tablet – next generation sequence assembly visualization. Bioinformatics. 2010;26:401–402. doi: 10.1093/bioinformatics/btp666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carver T, Bohme U, Otto TD, Parkhill J, Berriman M. BamView: viewing mapped read alignment data in the context of the reference sequence. Bioinformatics. 2010;26:676–677. doi: 10.1093/bioinformatics/btq010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Qi J, Zhao F, Buboltz A, Schuster SC. inGAP: an integrated next-generation genome analysis pipeline. Bioinformatics. 2010;26:127–129. doi: 10.1093/bioinformatics/btp615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Medvedev P, Stanciu M, Brudno M. Computational methods for discovering structural variation with next-generation sequencing. Nat. Methods. 2009;6:S13–S20. doi: 10.1038/nmeth.1374. [DOI] [PubMed] [Google Scholar]
- 14.Rozen S, Skaletsky H. Primer3 on the WWW for general users and for biologist programmers. Methods Mol. Biol. 2000;132:365–386. doi: 10.1385/1-59259-192-2:365. [DOI] [PubMed] [Google Scholar]
- 15.Li H, Ruan J, Durbin R. Mapping short DNA sequencing reads and calling variants using mapping quality scores. Genome Res. 2008;18:1851–1858. doi: 10.1101/gr.078212.108. [DOI] [PMC free article] [PubMed] [Google Scholar]