Abstract
The estimation of genetic linkage maps is a key component in plant and animal research, providing both an indication of the genetic structure of an organism and a mechanism for identifying candidate genes associated with traits of interest. Because of this importance, several computational solutions to genetic map estimation exist, mostly implemented as stand-alone software packages. However, the estimation process is often largely hidden from the user. Consequently, problems such as a program crashing may occur that leave a user baffled. THREaD Mapper Studio (http://cbr.jic.ac.uk/threadmapper) is a new web site that implements a novel, visual and interactive method for the estimation of genetic linkage maps from DNA markers. The rationale behind the web site is to make the estimation process as transparent and robust as possible, while also allowing users to use their expert knowledge during analysis. Indeed, the 3D visual nature of the tool allows users to spot features in a data set, such as outlying markers and potential structural rearrangements that could cause problems with the estimation procedure and to account for them in their analysis. Furthermore, THREaD Mapper Studio facilitates the visual comparison of genetic map solutions from third party software, aiding users in developing robust solutions for their data sets.
INTRODUCTION
The estimation of genetic maps is an important process in biological research. For organisms lacking genome sequences, genetic maps provide both an essential resource to understand the order and spacing of DNA markers and a reference for comparison with the genetic maps and genomes sequences of other organisms. In plant and animal breeding studies, genetic maps underpin the further analysis of key genes, including quantitative trait loci (QTL). For organisms whose genomes have been sequenced, genetic maps enable a bridge to the corresponding physical maps to be constructed, facilitating the identification of candidate genes implicated in key QTL and promoting improvement programs, should these be required, via marker-assisted selection methods.
Stated basically, for a data set of m markers scored for n individuals derived via a mapping experiment, the genetic mapping problem is firstly to divide the markers into l distinct linkage groups and secondly, for each of the linkage groups in turn, to order the markers along it and to find the distances between adjacent markers in the centiMorgan scale. It is now almost 100 years since the first genetic linkage map was developed by Sturtevant in 1913 (1). In this time, the genetic mapping problem has been studied widely (2–5) and several computational solutions have been proposed. Like any biological problem for which computational solutions readily exist, it is vital that users have access to robust and user-friendly software that allow them to analyse their data sets as quickly, easily and accurately as possible. Indeed, a number of software packages and web sites exist (6) implementing the various computational solutions and many have taken great care to optimize these valuable features. Despite the ease of use of software packages, fundamental problems with the estimation procedure remain. For example, software may fail for a given data set without apparent reason, or users may feel a lack of control over or understanding of their data set. Some of these problems may be attributed to specific features of the data sets themselves, such as outlying markers or chromosomal rearrangements between the parents of an experimental cross. Furthermore, these problems can sometimes be identified by visual analysis of the data sets, such that software with multiple visualization capabilities will offer the greatest opportunities for dealing with such problems before extensive computations are performed. In addition, different software packages may provide different solutions for a given data set, in which case it is good practice to compare these different solutions and to use them appropriately to create a robust final solution.
The THREaD Mapper method was developed to enable the visual analysis and interpretation of genetic mapping data sets using a novel computational method, exploiting the ‘horseshoe effect’ commonly observed upon the principal co-ordinates analysis of linearly-derived data sets (7). In the first instance, the method was developed as an in-house tool for the analysis of pea data sets. Since then it has been used in the international collaborative analysis of a Brachypodium genetic map (8) and in in-house wheat data sets. Most recently, a public web server THREaD Mapper Studio has been developed to enable any user to analyse their data set using the THREaD Mapper method without the need for downloading and installing software packages. Several motivating factors have led to the rationale and functionality of the THREaD Mapper Studio web server. Firstly, we wished to provide a simple yet powerful analytical solution to the problem that was unlikely to fail for most data sets. Secondly, we wished to break down the analytical process into a series of steps, guiding users through their analyses. Thirdly, we wished to provide attractive, Windows-style interfaces that users would find comfortable using. Fourthly, we wished the software to be fully functional within a web browser, so that users would not usually need to download and install software in order to use it. Fifthly, we wanted to provide simple, visual ways of comparing solutions between different genetic mapping tools. Finally, and perhaps most importantly, we wanted to enable users to visualize their data sets in ways hitherto unavailable, promoting better understanding of their data sets and ultimately, more accurate genetic maps.
THE THREAD MAPPER STUDIO WEB SERVER
Computational approach
THREaD Mapper Studio uses a new algorithmic approach to estimate genetic maps. The rationale behind the approach is the ‘horseshoe effect’ observed following a principle co-ordinate analyses of a linearly-derived data set, when viewed on a 2D plot of the first two principal co-ordinates. Here, we exploit this effect, ‘threading’ a genetic map through the arch of marker data points. For the purposes of genetic map estimation, it is better to extend such an approach into 3D space, where such a plot becomes ‘snake-shaped’. This extension finds a natural ‘sweet spot’ at which the combination of user interaction and data fitting is optimized. The THREaD Mapper method uses a mixture of existing and bespoke algorithms to transform data from a mapping experiment into such a 3D plot and then to extract from the plot in a robust manner an estimate of a genetic map. A further series of algorithms based around the spectral embedding approach enable a data set consisting of multiple linkage groups to be visualized and analysed, also in 3D space. Full details of the algorithmic approach will be given in a forthcoming publication. However, outline details of each of the main algorithmic steps may be found in the Supplementary Data. The THREaD Mapper Studio web server implements this new method, breaking down the various tasks and user-led choices into a series of simple and visually attractive steps. Particularly valuable features inherent to the method means users can: (i) visualize their maps in 3D, often allowing them to spot errors or problems with their maps, (ii) compare their maps to those genetic maps estimated under alternative software methods or to physical maps, and (iii) use a combination of algorithm- and user-based decisions in developing their maps. Below, we will describe each of the steps required to perform an analysis within the THREaD Mapper Studio web server.
Data loading
The first step in a THREaD Mapper Studio analysis is to load a mapping data set into the web server. Users enter an n × m marker segregation data file (i.e. n individuals each scored for m markers), with four input formats permitted: the Locus file (.loc) format used by software such as JoinMap (9) and CarthaGène (10,11), the Raw file (.raw) format used by MapMaker (12), and two simple comma- or tab-separated values (CSV/TSV) formats that can be created with widely-used tools such as Microsoft Excel. Descriptions of these formats are provided in the ‘Documentation’ section on the front page of the web site. In addition, a small selection of real and synthetic data sets are provided, such that a single click will either download the data set for perusal or will take users immediately to the start of an analysis [e.g. the data set in Figure 1 of Cheema and Dicks (6) is provided as the ‘BiB data set with Header Attributes’]. Users are also asked to indicate the experimental design of the mapping experiment, with F2, Doubled Haploid (DH), Backcross (BC) and Recombinant Inbred Line (RIL), the currently available choices.
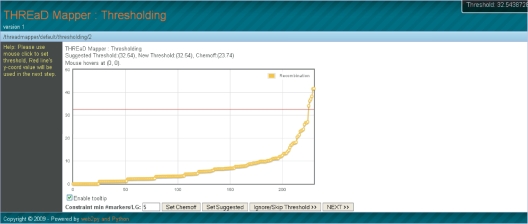
Figure 1.
A Thresholding plot for a barley data set of 232 markers. The horizontal red line shows the threshold chosen as the inter-marker distance beyond which to separate distinct linkage groups. In this case, this threshold cuts the data set into seven groups.
Data set refinement and parameter choice
The next part of the analysis is to choose the various analytical options for the data set undergoing analysis. This is carried out by a series of web pages that together simplify this process. To begin, once data loading has been successful, users are provided with simple statistics of their data set, such as P-values of segregation distortion for each marker, a pie-chart plot of the overall genotype frequencies for the data set and the number of missing scores for each marker. Based on these values, users may then choose to refine their data set by removing one or more markers from the analysis (e.g. markers with a large number of missing scores may prove unreliable in the estimation procedure). Users are then prompted to choose from a list a distance measure for inter-marker distance calculation. The various options given are tailored for the experimental design specified by the user and a default method is selected if a user is unsure which one to choose. Once pairwise distances have been calculated using the chosen scoring scheme, users are presented with a ‘heat-map’ of their data set, giving an indication of its overall linkage structure. Users are then asked whether their data set consists of a single or multiple linkage groups. At this stage, they are also asked whether they wish to generate an ensemble for the remainder of the analysis. The ensemble procedure perturbs the data sets in a carefully controlled manner to see how robust the resulting maps are to local error, with a growing value of the ensemble parameter T corresponding to a stronger perturbation. Users not wishing to carry out an ensemble analysis can simply choose the default value of T = 1 (no ensemble).
Thresholding
The next step of the analysis, known as ‘thresholding’, calculates a minimum spanning tree (MST) from the distance matrix and plots the sizes of pairwise inter-marker distances between markers adjacent in the MST, with values ordered according to size (i.e. smallest inter-marker distances to the left of the plot and highest to the right). Depending on the data set, such a plot may or may not indicate that more than one linkage group exists for the data set. In general, where such a plot exhibits a ‘jump’ between the majority of small distances and the minority of larger distances, there is good evidence for more than one linkage group. In such a case, large distances tend to represent those between linkage groups and small distances those within linkage groups and in practice, we would like to partition the data set into linkage groups accordingly. The Thresholding web page permits three ways to partition a behind-the-scenes graphical structure of the user's data set, in addition to allowing partitioning to be ignored. Firstly, the user may choose their own distance value threshold beyond which connections between pairs of markers are severed, by sliding up or down the red horizontal bar on the plot. Secondly, the theoretically-based Chernoff bound may be used, as introduced in the MSTMap method (13). Thirdly, an empirically-based value calculated by THREaD Mapper may be chosen. Figure 1 shows an example of empirically-based thresholding, for the Oregon Wolfe Barley (OWB) barley DH population with 232 markers and 94 individuals data set, downloaded from http://barleyworld.org/oregonwolfe.php on February 11, 2008. The plot shows a clear jump between the 224th and 225th highest inter-marker distances within the data set, representing the shift from within-linkage group to between-linkage group distances. Once the thresholding step has been completed, the user is presented with a spreadsheet-like screen showing the marker membership of each group (with a single grouping if no cutting has been performed).
Embedding the data set
The next part of the analysis is known as the Embedding, where the 3D transformation of a user's data set is calculated and displayed. First, the user is asked to choose from one of four embedding methods, with multi-dimensional scaling suitable for single-linkage groups and Spectral Embedding for multiple-linkage groups. The Isomap and Robust Kernel Isomapping methods are also applicable to data sets with multiple linkage groups but their performance makes them suitable for smaller data sets. Once this final choice has been made, THREaD Mapper Studio displays the main Embedding Screen. The analysis carried out within it is largely dependent on whether the data set derives from a single linkage group or from multiple linkage groups. For the former, the embedded markers are represented as a central, spinnable 3D snake-shaped plot, with markers represented by spheres that are projected onto a ‘trendline’ or ‘thread’ running through the centre of the spheres. For data sets consisting of multiple linkage groups, the embedded markers are represented by a single and more complex 3D graphical structure. Again, markers are represented by spheres with the colour of links between markers dependent on the strength of evidence that they are genetically linked. Crucially, marker spheres may be coloured in two ways. Firstly, they may be coloured according to user-input known as Attribute Groups where, for example, the results of a third-party linkage analysis may be encoded allowing it to be compared directly with the THREaD Mapper output. Alternatively, markers may be coloured according to marker type or perhaps to the chromosomal assignment of orthologues in a second genome. Secondly, markers may be coloured according to the estimated linkage group structure, perhaps determined via the Thresholding step.
The overall topology of the embedded 3D marker structure, the colouring provided by the Attribute Groups and other knowledge possessed by the user guide the final division of the markers into their respective linkage groups. This division, perhaps just a fine-tuning of a Thresholding partitioning, is achieved by ‘cutting’ and ‘joining’ pairs of markers, using either the buttons on the uppermost menu bar or the pairwise connections in the right hand panel. Once a final linkage group designation has been completed, each group may then in turn undergo individual analysis, as described below, to create a marker order and associated inter-marker distances. Figure 2 shows a Spectral Embedding of the OWB data set, with inter-marker distances calculated according to the Kosambi map function (14), where the seven linkage groups may easily be seen.
Figure 2.
A Spectral Embedding plot of the barley data set described in the text. The seven linkage groups detected in the Thresholding step are clearly spatially distinct within this plot. Other panels within the plot show marker membership of each linkage group and details of adjacent markers and their inter-marker distances.
Creating a marker order
Once a linkage group consisting of three or more markers has been selected for ordering and spacing, it is analysed using a novel algorithmic approach called non-linear geodesic smoothing. Here, a trendline is ‘threaded’ through the snake-shaped curve of the markers plotted in 3D space, an approach that gives the method its name. The marker ordering web page contains functions to produce the main THREaD Mapper output of the resulting genetic maps in various formats, including as a CSV file of marker distances, a PDF file containing a ‘cartoon’ of a linear genetic map and a pair of graphical heat-maps pre- and post-analysis.
Spotting unusual features
Every data set is different and may be affected by experimental error in one or more of a number of ways, and to differing degrees. One of the strengths of THREaD Mapper Studio is the manner in which it allows users to visualize a data set in 3D space without making restrictive assumptions about the way the data were derived. This means that the data are not forced to take on a certain structure but rather find their own structure that can be interpreted appropriately. For example, in the embedding procedure marker points are not forced onto the trendline running through them. Instead, projection lines are simply joined from the markers onto the trendline. This means that markers that lie remote from the trendline may be seen easily and users can make choices about their reliability and their affect on the analysis (and indeed such markers may then be removed if wished).
Help for the user
Help for users is provided in three ways. Firstly a PDF help document may be downloaded from the ‘Documentation’ section on the THREaD Mapper Studio homepage. The help document describes the individual parts of an analysis, discussing the implications of choices a user may make, and runs through a basic analysis on one of the example files provided. Secondly a tutorial, which requires Flash functionality within a user's web browser, is accessed via the ‘Tutorials’ section on the homepage. This tutorial emulates a typical analysis, showing the series of mouse moves and choices that enables the analysis to be performed. Thirdly, brief help tips are provided on the web pages themselves, tailored to the particular analytical step in hand.
Technical features
THREaD Mapper Studio is mostly coded in Python and JavaScript. It runs on the web2pyTMEnterprise Web Framework, an open source full-stack enterprise framework for agile development of portable database-driven applications that is written in Python (http://www.web2py.com/). The THREaD Mapper Studio GUI is developed using ExtJS version 2.4 (http://www.extjs.com/), a JavaScript platform for development of cross-browser web applications. Ajax capabilities are supported using the jQuery Javascript library (http://jquery.com/) and plotting is supported by Flot (http://code.google.com/p/flot/) and the Google Chart API (http://code.google.com/apis/charttools/). 3D visualization of the embedded marker data sets is achieved using Java Applet Jmol version 11.8 (http://jmol.sourceforge.net/). Graph/network creation, manipulation and analysis are supported using the NetworkX Python package (http://networkx.lanl.gov/) and numerical Python modules such as numpy/scipy (http://numpy.scipy.org/). The ‘Acknowledgements’ section on the THREaD Mapper Studio homepage provides a complete list of the various Python modules used.
Requirements
Use of the THREaD Mapper Studio web server does not require users to download the THREaD Mapper software onto their computers. However, certain web browser plugins are necessary for the software to function fully. In particular, THREaD Mapper Studio requires that a user's web browser is both JavaScript and Java Runtime Environment enabled. Furthermore, the Adobe Flash Player plugin is required in order to view the Tutorial screencast mounted on the THREaD Mapper Studio homepage. We have tested the web site extensively within the Internet Explorer 7, Internet Explorer 8, Firefox and Google Chrome browsers under Windows XP.
CONCLUSION
THREaD Mapper Studio is a new web server presenting a novel way to construct and compare genetic maps. In developing it we have attempted to overcome two of the major issues with software for genetic map estimation: software failures and difficulties in controlling or understanding a data set during analysis. Wherever possible, we have attempted to keep our algorithms as simple as possible, contributing to its robustness. We have allowed users the opportunity to override or edit computational results, so that they are ultimately in control of an analysis and its solution. Throughout an analysis, we have provided multiple, interactive data visualizations so that users can understand their data sets better. Indeed, for many complex data sets such as mapping data, visualization can be key to understanding many of its features. The 3D and visual nature of the THREaD Mapper method give it a natural affinity with user-interaction and so this feature has been optimized throughout. However, such visual attractiveness and ease of use should not come at the expense of performance and we have endeavoured to strike a balance between the user interface and the power of the algorithms. At present, following testing with multiple real and simulated data sets (see Supplementary Data for details of benchmark analyses), we believe the web server to be suitable for the analysis of data sets of up to ∼1000–1500 markers, depending on the complexity of the data set. Indeed, the web server currently limits an analysis to data sets of up to 2000 unique markers and an execution time of each algorithmic step to 15–20 min, depending on the step. However, we know that high-throughput marker technologies are now capable of developing data sets with tens of thousands of markers. Consequently, in the near future we will look to extend the method, in tandem with the web server, for the analysis of high-throughput molecular marker data sets. In addition, we have plans to release a stand-alone version of the THREaD Mapper software, enabling users to analyse data sets on their own computing resources.
FUNDING
European Union Framework VI Grain Legumes Integrated Project (FOOD-CT-2004-506223); Biotechnology and Biological Sciences Research Council (John Innes Centre Core Strategic Grant). Funding for open access charge: Biotechnology and Biological Sciences Research Council.
Conflict of interest statement. None declared.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
Many of our colleagues and collaborators have given valuable guidance on the practicalities of genetic map estimation, help with data sets and ideas for improving the THREaD Mapper Studio web site, and we would like to thank them all. Particular thanks are due to David Garvin, John Snape and Luzie Wingen. We would also like to thank two anonymous referees for their constructive criticism of this manuscript and for their helpful suggestions for improving the THREaD Mapper Studio web server.
REFERENCES
- 1.Sturtevant AH. The linear arrangement of six sex-linked factors in Drosophila, as shown by their mode of association. J. Exp. Zool. 1913;14:43–59. [Google Scholar]
- 2.Edwards AWF. Linkage methods in human genetics before the computer. Human Genet. 2005;118:515–530. doi: 10.1007/s00439-005-0049-1. [DOI] [PubMed] [Google Scholar]
- 3.Chakravarti A, Lynn A. Meiotic mapping in humans. In: Birren B, Green ED, Hieter P, Klapholz S, Myers RM, Riethman H, Roskams J, editors. Genome Analysis: A Laboratory Manual Series. Volume 4:Mapping Genomes. Plainview NY: Cold Spring Harbor Laboratory Press; 1999. pp. 1–70. [Google Scholar]
- 4.Nelson JC. Methods and software for genetic mapping. In: Meksem K, Kahl G, editors. The Handbook of Plant Genome Mapping. Wiley-VCH Verlag GmbH & Co. KGaA; 2005. pp. 53–74. [Google Scholar]
- 5.Semagn K, Bjørnstad Å, Ndjiondjop MN. Principles, requirements and prospects of genetic mapping in plants. Afr. J. Biotechnol. 2006;5:2569–2587. [Google Scholar]
- 6.Cheema J, Dicks J. Computational approaches and software tools for genetic linkage map estimation in plants. Brief. Bioinformatics. 2009;10:595–608. doi: 10.1093/bib/bbp045. [DOI] [PubMed] [Google Scholar]
- 7.Podani J, Miklos I. Resemblance coefficients and the horseshoe effect in principal coordinates analysis. Ecology. 2002;83:3331–3343. [Google Scholar]
- 8.Garvin DF, Mckenzie N, Vogel JP, Mockler TC, Blankenheim ZJ, Wright J, Cheema JJS, Dicks J, Huo N, Hayden DM, et al. An SSR-based genetic linkage map of the model grass Brachypodium distachyon. Genome. 2010;53:1–13. doi: 10.1139/g09-079. [DOI] [PubMed] [Google Scholar]
- 9.Stam P. Construction of integrated genetic linkage maps by means of a new computer package: JoinMap. Plant J. 1993;3:739–744. [Google Scholar]
- 10.Schiex T, Gaspin C. CarthaGène: constructing and joining maximum likelihood genetic maps. Proceedings of the International Conference on Intelligent Systems for Molecular Biology 1997. 1997;Abstract 5:258–267. [PubMed] [Google Scholar]
- 11.de Givry S, Bouchez M, Chabrier P, Milan D, Schiex T. CarthaGène: multipopulation integrated genetic and radiation hybrid mapping. Bioinformatics. 2005;21:1703–1704. doi: 10.1093/bioinformatics/bti222. [DOI] [PubMed] [Google Scholar]
- 12.Lander ES, Green P, Abrahamson J, Barlow A, Daly MJ, Lincoln SE, Newberg LA. MAPMAKER: an interactive computer package for constructing primary genetic linkage maps of experimental and natural populations. Genomics. 1987;1:174–181. doi: 10.1016/0888-7543(87)90010-3. [DOI] [PubMed] [Google Scholar]
- 13.Wu Y, Bhat PR, Close TJ, Lonardi S. Efficient and accurate construction of genetic linkage maps from the minimum spanning tree of a graph. PLoS Genet. 2008;4:e1000212. doi: 10.1371/journal.pgen.1000212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kosambi DD. The estimation of map distances from recombination values. Ann. Eugen. 1944;12:172–175. [Google Scholar]