Abstract
R spider is a web-based tool for the analysis of a gene list using the systematic knowledge of core pathways and reactions in human biology accumulated in the Reactome and KEGG databases. R spider implements a network-based statistical framework, which provides a global understanding of gene relations in the supplied gene list, and fully exploits the Reactome and KEGG knowledge bases. R spider provides a user-friendly dialog-driven web interface for several model organisms and supports most available gene identifiers. R spider is freely available at http://mips.helmholtz-muenchen.de/proj/rspider.
INTRODUCTION
High-throughput technologies enable biological researchers to study hundreds or thousands of genes simultaneously. Genes or proteins are detected that are differentially expressed or co-expressed across varying cellular conditions. However, generating hypotheses about the underlying biological mechanisms based on experimentally derived gene/protein lists remains a non-trivial task for biologists. In 2002, a computerized analysis approach using the Gene Ontology (GO) was proposed to deal with this issue (1,2). Currently, there are over 25 tools performing this type of analysis with some variations (3–13). More recently, computational methods seek to interpret or at least visualize the pathway context of the experimentally derived genes (14–17). In this respect, one should mentioned a landmark procedure proposed recently in (17,18) which goes beyond gene pairs and fully captures the topology of signaling pathways by propagating the perturbations measured at gene levels through the entire pathway. However, the development of rigorous statistical methods for global network inference has been a challenging task.
Recently, we have introduced a network-based computational framework for the interpretation of gene/protein lists derived from high-throughput studies (19,20). Our approach overcomes a major bottleneck of the commonly employed methods for enrichment analysis (21) by providing network models that unite genes from different pathways into a single connected network. A Monte Carlo procedure was employed to estimate the significance of the inferred models, thus providing a rigorous quantitative statistical control (22). A web-based tool, KEGG spider (19), was introduced that exploits the network-based methodology for the exploration of metabolic reactions accumulated in the KEGG database (23). It was demonstrated that KEGG spider provides deeper insight into the genomic basis of metabolism variations in comparison to other tools (19).
Although being a powerful tool, KEGG spider is limited only to metabolism-related genes which cover <10% of the human genome (about 1100 genes). It is clear that many other important cellular processes, such as regulatory and signaling pathways remain uncovered by the inferred network models. On the other hand, the Reactome knowledgebase (24,25) is a dynamically expanding project, which provides high quality expert-authored, peer-reviewed knowledge of human reactions and pathways, covering 3916 human proteins (as of release 30). To provide experimentalists with an efficient web-based tool for the analysis of high-throughput data using Reactome knowledge, we have developed R spider, which implements the network-based methodology and exploits the data accumulated in the Reactome knowledgebase to the full extent. R spider unites both Reactome and KEGG knowledge databases covering proteins from signaling and metabolism pathways.
We would like to point out that there are other signaling and metabolic databases available in the public domain like the manually curated BioCarta, NCI or inferred data (26) or (27). R spider has the option to switch between Reactome&KEGG, Nature Curated pathways (http://pid.nci.nih.gov/) and BioCarta (www.biocarta.com).
MATERIALS AND METHODS
A global Reactome protein network
Reactome (http://www.reactome.org/) is an expert-authored, peer-reviewed knowledgebase of human reactions and pathways. We used a file in tab-delimited format which specifies protein–protein interaction pairs derived from Reactome data (http://www.reactome.org/download/current/homo_sapiens.interactions.txt.gz). The meaning of ‘interaction’ is quite broad: two protein sequences occur in the same complex or they occur in the same or neighbouring reaction(s). For the human genome, the global Reactome protein network covers about 3700 proteins (including proteins from non-human species that interact with human proteins) involved in approximately 83 000 unique pairwise interactions (based on release 30).
A global metabolic gene network
The KEGG database is a collection of chemical structure transformation patterns for substrate–product pairs (reactant pairs). A detailed description of the procedure used to construct a global metabolic gene network can be found in ref. (19). The resulting global metabolic gene network links by edges any two genes that are associated with reactions sharing common compounds (from the main reaction pair). For the human genome, the global metabolic gene network covers about 1100 genes involved in approximately 15 000 unique pairwise interactions.
Integral reference network
To unite both networks, the Reactome protein network was transformed into a gene network. As in many cases, several proteins map to the same gene, the resulting gene network has a smaller number of nodes and edges. Once both KEGG and Reactome networks have the same type of node identifiers, they can be united. For the human genome, the resulting integral network covers about 3700 genes involved in approximately 50 000 unique pairwise gene interactions.
Network inference procedure and statistical treatment
Detailed information on the network inference and the Monte Carlo simulation procedure for computing P-values can be found in our previously published papers (19,20,28).
Initially, the genes from the input list are mapped to the global reference network. At this point, all nodes from the input list are disconnected. In the first step, all pairs of nodes with distance 1 are connected by edges and connected subnetworks are extracted. The subnetwork with the maximal number of nodes is referred to as an inferred network model D1. In the second step, the disconnected nodes from the input list with distance 2 are connected by edges. The subnetwork with the maximal number of input nodes is inferred and referred to as network model D2. In the next step, the disconnected nodes from the input list with distance 3 are connected by edges and a network model D3 (a subnetwork with the maximal number of input nodes) is inferred. Models D2 and D3 incorporate nodes that are not from the input list but are added to connect input nodes in the network model. We refer to these added nodes as intermediate or missing genes.
Let us assume that we have N genes from the input list to be mapped to the reference network. Next, we refer to the value N as the size of the input list. We infer the network models D1, D2, D3. Let us denote S1, S2, S3 to be the number of input nodes in the inferred network models. We also refer to S1, S2, S3 as the sizes of the respective models D1, D2, D3. Given the number of mapped genes from the input list (N), we consider the sizes of the models (values S1, S2, S3) as statistics. We have to estimate the probability to get models of the same or larger sizes from a randomly generated gene list which has N genes mapped to the reference network.
To generate the background distributions BD1, BD2, BD3 we repeat the following simulation procedure k times, where k specifies the upper significance level. A random gene list Lj of size N (equal to the size of the input list) is generated by sampling genes from global gene network. Index j = 1 … k specifies each of the k random simulations. The network inference procedure described above is applied to the random list Lj and the network models D1, D2, D3 are inferred. Let us denote the size (the number of input genes) of the inferred models D1, D2, D3 for the random list Lj as R1j, R2j, R3j. Thus, after repeating the simulation procedure k times, we get the background distribution R1j (j = 1… k) for models D1, the background distribution R2j (j = 1… k) for models D2, and the background distribution R3j (j = 1… k) for models D3.
To estimate significance of the inferred network model D1 for the input gene list, the value S1 is compared with the distribution R1j. Let n be the number of values from the distribution R1j that are equal or greater than S1. The estimate of P (P-value) of the inferred network model D1 is computed as P = (n + 1)/k. In the same way the P-values for the model D2 and D3 are estimated.
Statistical treatment plays an important role for the quality control of inferred models. It is clear that given a gene list and a reference network, one can always infer some model, which will cover all genes from the list by relaxing the number of possible intermediate genes. There is a very simple test for any similar tool: the tool must be able to recognize a random gene list and return on average insignificant P-values for the random case. In 20 submissions of different randomly generated gene lists on average only 1 case is expected to be significant at the level of 0.05 (1/20). The estimate of the P-value provided by the Monte Carlo procedure corresponds exactly to the definition of P-value: the probability to get a model of the same quality for a random gene list.
Enrichment of the reactome and KEGG canonical pathways
To compute enrichment of canonical Reactome and KEGG pathways, we also employed the Monte Carlo procedure. In this case, we randomly draw k genes (the number of genes in the input list) 100 times from the set of all genes (or from the background set of genes supplied by the user) and each time we estimate P-value based on the hypergeometric distribution for the best (whatever) pathway. Thus, we got a distribution of size 100 of the best P-values for a random drawing of k genes which we compare with the P-value for the best (whatever) pathway related to our original list. The estimate of the adjusted P-value by Monte Carlo procedure is given by the share of random simulations where the best P-value was equal or superior (less) than the P-value for the best (whatever) pathways related to our original gene list.
RESULTS
R spider (http://mips.helmholtz-muenchen.de/proj/rspider) is a freely available web-based tool that implements a pathway-free statistical framework for the interpretation of gene lists from high-throughput studies. R spider is available for several model organisms (Mus musculus, Rattus norvegicus, Caenorhabditis elegans, Saccharomyces cerevisiae, Arabidopsis thaliana, Drosophila melanogaster). In addition, R spider has the option to switch to the other available in the public domain signaling pathway databases, Nature Curated pathways (http://pid.nci.nih.gov/) and BioCarta (www.biocarta.com).
R spider has a simple, user-friendly interface. As input it accepts several types of gene or protein identifiers, such as identifiers from ‘Entrez Gene’ (29), ‘UniProt/Swiss-Prot’ (30), ‘Hugo Gene Symbols’, ‘UniGene’, ‘Ensembl’ (31), ‘RefSeq’ (32) and ‘Affymetrix’ (33). As output, the user obtains network models (D1, D2, D3), where (1, 2, 3) indicates the maximal distance between any two input genes to be considered as ‘connected’ in the output model. The network model (D1, D2 or D3) represent a connected subnetwork with the maximal number of input genes. R spider provides a report on the statistical significance of the inferred network models (D1, D2, D3), as well as a catalog of the enriched Reactome or KEGG pathways. For each model (D1, D2, D3), a link is provided to obtain a graphical visualization. The visualization is performed by the Medusa package (34). We would like to point out that online visualization capabilities are limited. For this reason, we recommend to download the inferred network models as text files (links are provided on the visualization page) and to use freely available packages (Cytoscape, Meduza) for network visualization. Using these programs the users can produce high-quality figures (34,35).
Graphical output
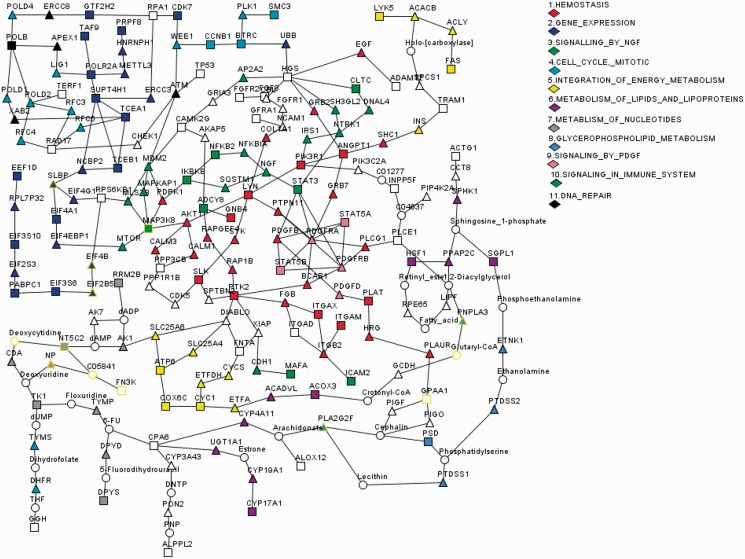
In the graphical output, input genes are represented by rectangles and specified by the input gene Ids. Intermediate genes are represented by triangles and specified by Entrez Gene Symbols. Compounds are represented by circles and specified by compound names (if the length of the name exceeds 10 digits then the compound KEGG id is used). Different colors are used to specify canonical Reactome or KEGG pathways. In general, up to 11 of the most representative pathways (in terms of the number of genes in the model, both input and intermediate genes are counted) are coloured. In most cases, a gene can be associated to multiple pathways. For this reason, R spider implements a strict hierarchical procedure for gene coloring. First, pathways are ordered in respect to the number of genes that are present in the model from any given pathway. The most representative pathway will be colored in red. Colored genes (red) are excluded and pathways are reordered considering only the remaining genes. The next most represented pathway will be colored in blue. Colored genes (red and blue) are excluded and pathways are reordered considering only the remaining genes. The procedure will continue until 13 pathways will be colored or there will be no pathway which covers at least two genes. Therefore, colors have a strict hierarchy: red, blue, green and so on. The number before the color indicates the hierarchy order (Figure 1). It is evident that some red genes may also belong to the blue (green and so on) pathway, but not vice versa.
Figure 1.
Network model D3 returned by R spider on submission of 360 candidate genes residing in regions with copy number alteration typical of the Sézary syndrome (37). Boxes represent input genes, triangles represent intermediate genes (genes that are added to connect two input genes, for model D3 up to two intermediate genes are allowed between any two input genes), circles represent compounds which are common substrates or products for both connected genes. Diamonds are used to specify the colour of canonical Reactome or KEGG pathways.
Table: interaction context
For each gene in the reported model, R spider provides the full interaction context. This information is summarized in the table ‘Interacting Pairs’. In the case of Reactome, there are four types of interactions: ‘direct_complex’, ‘indirect_complex’, ‘reaction’ or ‘neighbouring_reaction’. In the case of the KEGG database, interactions represent either a compound (connected genes are assigned to different reactions utilizing the same compound) or, rarely, by a reaction ID (both connected genes catalyze the same metabolic reaction). The edge can be supported by several different interactions, all of which will be reported, and corresponding links to the source data are provided.
Example
We present at our website (http://mips.helmholtz-muenchen.de/proj/rspider/example.html) several hundred examples of analyses by R spider of gene lists, which were automatically extracted by text mining from proteomics studies in various biological contexts (36). Here, we present one example in detail to demonstrate the potential benefit of our tool.
Currently, many clinical studies are designed to reveal possible pathogenic mechanisms and novel therapeutic targets for complex diseases with specific phenotypes. The Sézary syndrome, for example, is associated with the aggressive cutaneous T-cell lymphoma/leukemia. In a study by Vermeer et al. (37), a high-resolution array-based comparative genomic hybridization was performed on malignant T cells from 20 patients to reveal highly recurrent genetic alterations typical for the Sézary syndrome. Minimal common regions with copy number alteration occurring in at least 35% of patients were reported, which comprised in total about 360 candidate genes (see Table 1 in ref. 37).
Only 22 of these genes are mapped to KEGG metabolic pathways. Thus, for comparison, an analysis by KEGG spider reports that the inferred network model is not significant (P = ∼0.1). On the contrary, consideration of the integral reference network that unites both Reactome and KEGG data provides more interesting insights into the possible molecular mechanisms behind genes with copy number alteration in the Sézary syndrome. In this case, 92 out of the 360 genes are mapped to the integral network. Network model D3, which allows up to two missing genes between any two input genes, connects 74 out of the 92 mapped candidate genes into a single non-interrupted network. The model is statistically significant (P < 0.01). R spider randomly sampled 92 genes from the set of 3700 human genes that constitute the integral reference network for 1000 times; and in 993 cases, the size of the resulting network model D3 was less than 74 genes. Thus, the significance of the model is about 0.01.
R spider provides graphical models. The network model D3 for the considered example, which covers 74 genes (P < 0.01), is presented in Figure 1. Proteins from the input list are indicated by rectangles, intermediate proteins by triangles, and chemical compounds are indicated by circles. The colours are used to specify Reactome and KEGG canonical pathways.
In comparison to other available pathway analyses tools, R spider provides a global vision of gene functional relations. For example, submission to Onto-express (17) results in reporting of several (∼10) enriched pathways with possibility to visualize them separately. This is certainly valuable information. However, the best model (enriched pathway ‘Pathways in cancer’) covers 19 genes. The relation between pathways as well as the role and relation between genes that are not covered by enriched pathways is not disclosed. Thus, in comparison to Onto-express R spider demonstrates that genes residing in regions which frequently have a copy number alteration in Sézary syndrome are dependent although they belongs to a wide spectrum of signaling and metabolic pathways. In this case the user gets a newly created pathway which covers 74 genes and actually runs through several canonical Reactome and KEGG pathways.
CONCLUSIONS
Various modern genomics technologies result in gene lists. A better understanding of the biological mechanisms, which unite the identified genes, can give clues to a better understanding of the phenomena under study. R spider provides a possibility to actively exploit the knowledge of biological processes of various natures accumulated in the Reactome knowledgebase and metabolism related processes in the KEGG database to decipher the mechanisms behind experimentally derived gene lists. A pathway-free statistical framework combined with the most advanced publicly available databases for pathways and reactions makes R spider a very attractive tool for interpretation of genomics data.
FUNDING
Funding for open access charge: European Bioinformatics Institute, Wellcome Trust Genome Campus; The development of Reactome is supported by a grant from the US National Institutes of Health (P41 HG003751) and EU grant LSHG-CT-2005-518254 “ENFIN”.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We thank Philip Wong for helpful discussions.
REFERENCES
- 1.Draghici S, Khatri P, Martins RP, Ostermeier GC, Krawetz SA. Global functional profiling of gene expression. Genomics. 2003;81:98–104. doi: 10.1016/s0888-7543(02)00021-6. [DOI] [PubMed] [Google Scholar]
- 2.Khatri P, Draghici S, Ostermeier GC, Krawetz SA. Profiling gene expression using onto-express. Genomics. 2002;79:266–270. doi: 10.1006/geno.2002.6698. [DOI] [PubMed] [Google Scholar]
- 3.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl Acad. Sci. USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Reimand J, Kull M, Peterson H, Hansen J, Vilo J. g:Profiler—a web-based toolset for functional profiling of gene lists from large-scale experiments. Nucleic Acids Res. 2007;35:W193–W200. doi: 10.1093/nar/gkm226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Masseroli M, Martucci D, Pinciroli F. GFINDer: Genome Function INtegrated Discoverer through dynamic annotation, statistical analysis, and mining. Nucleic Acids Res. 2004;32:W293–W300. doi: 10.1093/nar/gkh432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Martin D, Brun C, Remy E, Mouren P, Thieffry D, Jacq B. GOToolBox: functional analysis of gene datasets based on Gene Ontology. Genome Biol. 2004;5:R101. doi: 10.1186/gb-2004-5-12-r101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Khatri P, Voichita C, Kattan K, Ansari N, Khatri A, Georgescu C, Tarca AL, Draghici S. Onto-Tools: new additions and improvements in 2006. Nucleic Acids Res. 2007;35:W206–W211. doi: 10.1093/nar/gkm327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dietmann S, Georgii E, Antonov A, Tsuda K, Mewes HW. The DICS repository: module-assisted analysis of disease-related gene lists. Bioinformatics. 2009;25:830–831. doi: 10.1093/bioinformatics/btp055. [DOI] [PubMed] [Google Scholar]
- 9.Berriz GF, King OD, Bryant B, Sander C, Roth FP. Characterizing gene sets with FuncAssociate. Bioinformatics. 2003;19:2502–2504. doi: 10.1093/bioinformatics/btg363. [DOI] [PubMed] [Google Scholar]
- 10.Antonov AV, Schmidt T, Wang Y, Mewes HW. ProfCom: a web tool for profiling the complex functionality of gene groups identified from high-throughput data. Nucleic Acids Res. 2008;36:W347–W351. doi: 10.1093/nar/gkn239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Antonov AV, Dietmann S, Wong P, Lutter D, Mewes HW. GeneSet2miRNA: finding the signature of cooperative miRNA activities in the gene lists. Nucleic Acids Res. 2009;37:W323–W328. doi: 10.1093/nar/gkp313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Khatri P, Bhavsar P, Bawa G, Draghici S. Onto-Tools: an ensemble of web-accessible, ontology-based tools for the functional design and interpretation of high-throughput gene expression experiments. Nucleic Acids Res. 2004;32:W449–W456. doi: 10.1093/nar/gkh409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Alexa A, Rahnenfuhrer J, Lengauer T. Improved scoring of functional groups from gene expression data by decorrelating GO graph structure 2. Bioinformatics. 2006;22:1600–1607. doi: 10.1093/bioinformatics/btl140. [DOI] [PubMed] [Google Scholar]
- 14.Adler P, Reimand J, Janes J, Kolde R, Peterson H, Vilo J. KEGGanim: pathway animations for high-throughput data. Bioinformatics. 2008;24:588–590. doi: 10.1093/bioinformatics/btm581. [DOI] [PubMed] [Google Scholar]
- 15.Reimand J, Tooming L, Peterson H, Adler P, Vilo J. GraphWeb: mining heterogeneous biological networks for gene modules with functional significance. Nucleic Acids Res. 2008;36:W347–W351. doi: 10.1093/nar/gkn230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Berger SI, Posner JM, Ma'ayan A. Genes2Networks: connecting lists of gene symbols using mammalian protein interactions databases. BMC Bioinformatics. 2007;8:372. doi: 10.1186/1471-2105-8-372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Draghici S, Khatri P, Tarca AL, Amin K, Done A, Voichita C, Georgescu C, Romero R. A systems biology approach for pathway level analysis. Genome Res. 2007;17:1537–1545. doi: 10.1101/gr.6202607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tarca AL, Draghici S, Khatri P, Hassan SS, Mittal P, Kim JS, Kim CJ, Kusanovic JP, Romero R. A novel signaling pathway impact analysis 1. Bioinformatics. 2009;25:75–82. doi: 10.1093/bioinformatics/btn577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Antonov AV, Dietmann S, Mewes HW. KEGG spider: interpretation of genomics data in the context of the global gene metabolic network. Genome Biol. 2008;9:R179. doi: 10.1186/gb-2008-9-12-r179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Antonov AV, Dietmann S, Rodchenkov I, Mewes HW. PPI spider: a tool for the interpretation of proteomics data in the context of protein-protein interaction networks. Proteomics. 2009;9:2740–2749. doi: 10.1002/pmic.200800612. [DOI] [PubMed] [Google Scholar]
- 21.Khatri P, Draghici S. Ontological analysis of gene expression data: current tools, limitations, and open problems. Bioinformatics. 2005;21:3587–3595. doi: 10.1093/bioinformatics/bti565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Westfall PN, Young SS. Resampling-based Multiple Testing: Examples and Methods for P-value Adjustment. New York: John Wiley & Sons, Inc; 1993. [Google Scholar]
- 23.Ogata H, Goto S, Sato K, Fujibuchi W, Bono H, Kanehisa M. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 1999;27:29–34. doi: 10.1093/nar/27.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Matthews L, Gopinath G, Gillespie M, Caudy M, Croft D, de BB, Garapati P, Hemish J, Hermjakob H, Jassal B, et al. Reactome knowledgebase of human biological pathways and processes. Nucleic Acids Res. 2009;37:D619–D622. doi: 10.1093/nar/gkn863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vastrik I, D'Eustachio P, Schmidt E, Gopinath G, Croft D, de BB, Gillespie M, Jassal B, Lewis S, Matthews L, et al. Reactome: a knowledge base of biologic pathways and processes. Genome Biol. 2007;8:R39. doi: 10.1186/gb-2007-8-3-r39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kitano H, Oda K. Robustness trade-offs and host-microbial symbiosis in the immune system4. Mol. Syst. Biol. 2006;2:2006. doi: 10.1038/msb4100039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ma'ayan A, Jenkins SL, Neves S, Hasseldine A, Grace E, Dubin-Thaler B, Eungdamrong NJ, Weng G, Ram PT, Rice JJ, et al. Formation of regulatory patterns during signal propagation in a Mammalian cellular network16. Science. 2005;309:1078–1083. doi: 10.1126/science.1108876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Antonov AV, Mewes HW. BIOREL: the benchmark resource to estimate the relevance of the gene networks. FEBS Lett. 2006;580:844–848. doi: 10.1016/j.febslet.2005.12.101. [DOI] [PubMed] [Google Scholar]
- 29.Wheeler DL, Barrett T, Benson DA, Bryant SH, Canese K, Chetvernin V, Church DM, DiCuccio M, Edgar R, Federhen S, et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2006;34:D173–D180. doi: 10.1093/nar/gkj158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Boutet E, Lieberherr D, Tognolli M, Schneider M, Bairoch A. UniProtKB/Swiss-Prot. Methods Mol. Biol. 2007;406:89–112. doi: 10.1007/978-1-59745-535-0_4. [DOI] [PubMed] [Google Scholar]
- 31.Hubbard TJ, Aken BL, Beal K, Ballester B, Caccamo M, Chen Y, Clarke L, Coates G, Cunningham F, Cutts T, et al. Ensembl 2007. Nucleic Acids Res. 2007;35:D610–D617. doi: 10.1093/nar/gkl996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pruitt KD, Tatusova T, Maglott DR. NCBI reference sequences (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 2007;35:D61–D65. doi: 10.1093/nar/gkl842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu G, Loraine AE, Shigeta R, Cline M, Cheng J, Valmeekam V, Sun S, Kulp D, Siani-Rose MA. NetAffx: Affymetrix probesets and annotations. Nucleic Acids Res. 2003;31:82–86. doi: 10.1093/nar/gkg121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hooper SD, Bork P. Medusa: a simple tool for interaction graph analysis. Bioinformatics. 2005;21:4432–4433. doi: 10.1093/bioinformatics/bti696. [DOI] [PubMed] [Google Scholar]
- 35.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Antonov AV, Dietmann S, Wong P, Igor R, Mewes HW. PLIPS, an automatically collected database of protein lists reported by proteomics studies. J. Proteome. Res. 2009;8:1193–1197. doi: 10.1021/pr800804d. [DOI] [PubMed] [Google Scholar]
- 37.Vermeer MH, van Doorn R, Dijkman R, Mao X, Whittaker S, van Voorst Varder PC, Gerritsen MJ, Geerts ML, Gellrich S, Soderberg O, et al. Novel and highly recurrent chromosomal alterations in Sezary syndrome. Cancer Res. 2008;68:2689–2698. doi: 10.1158/0008-5472.CAN-07-6398. [DOI] [PubMed] [Google Scholar]