Abstract
GeneMANIA (http://www.genemania.org) is a flexible, user-friendly web interface for generating hypotheses about gene function, analyzing gene lists and prioritizing genes for functional assays. Given a query list, GeneMANIA extends the list with functionally similar genes that it identifies using available genomics and proteomics data. GeneMANIA also reports weights that indicate the predictive value of each selected data set for the query. Six organisms are currently supported (Arabidopsis thaliana, Caenorhabditis elegans, Drosophila melanogaster, Mus musculus, Homo sapiens and Saccharomyces cerevisiae) and hundreds of data sets have been collected from GEO, BioGRID, Pathway Commons and I2D, as well as organism-specific functional genomics data sets. Users can select arbitrary subsets of the data sets associated with an organism to perform their analyses and can upload their own data sets to analyze. The GeneMANIA algorithm performs as well or better than other gene function prediction methods on yeast and mouse benchmarks. The high accuracy of the GeneMANIA prediction algorithm, an intuitive user interface and large database make GeneMANIA a useful tool for any biologist.
INTRODUCTION
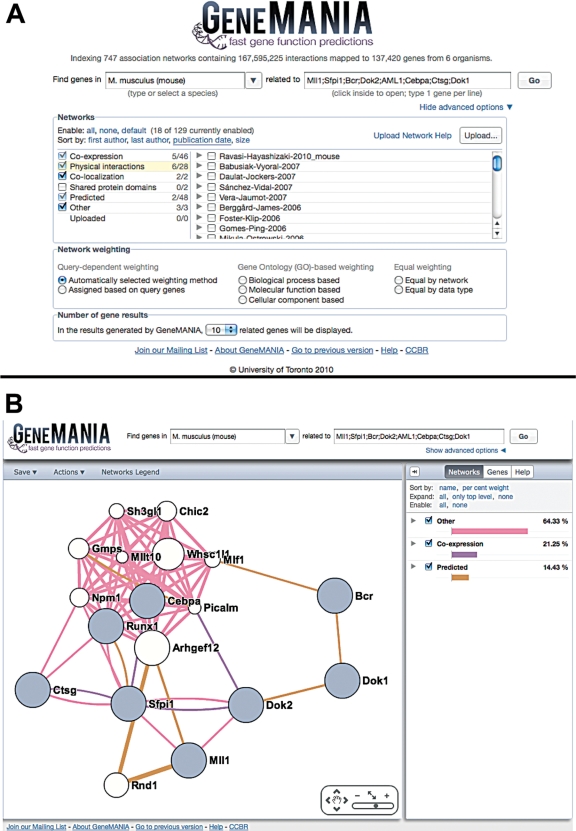
The input to GeneMANIA is simple—the user enters a list of genes and, optionally, selects from a list of data sets that they wish to query (Figure 1A). GeneMANIA then extends the user’s list with genes that are functionally similar, or have shared properties with the initial query genes, and displays an interactive functional association network, illustrating the relationships among the genes and data sets. For example, if the user enters protein complex members, such as yeast ARP2 and ARP3, GeneMANIA will output other complex components, and highly weight co-expression and protein interaction data sets. If the query genes are involved in disease, such as a mouse leukemia model, OMIM and phenotype data sets may receive high weight and GeneMANIA will output genes that likely are involved in the same process (Figure 1B). Users interested in prioritizing genes for planning a functional screen can use GeneMANIA to return ranked lists of genes likely to share phenotypes with those in the query list based on GeneMANIA’s large and diverse data collection.
Figure 1.
Images from http://www.genemania.org. (A) The initial query screen, with the advanced options panel expanded, providing the user the ability to select desired networks, choose a network weighting method and the number of genes to return. (B) The results page for the mouse default query, which is a set of genes involved in leukemia. Users can examine information associated with each gene and network by expanding their entries on the corresponding panel. We include linkouts to model organism databases (FlyBase, WormBase, SGD and TAIR) and to the Arabidopsis resource BAR (19) that is useful to plant users.
Another helpful feature of GeneMANIA is that it assigns weights to data sets based on how useful they are for each query. Individual data sets are represented as networks, and in the basic algorithm, each network is assigned a weight primarily based on how well connected genes in the query list are to each other compared with their connectivity to non-query genes. However, GeneMANIA’s adaptive weighting methods also detect and down-weight redundant networks. This network weighting feature is particularly useful for determining how genes in a gene list are connected to one another, or for determining which types of functional genomic data are most useful to collect for finding more genes like those in the query list.
Organisms and identifiers
We currently support six organisms: yeast (Saccharomyces cerevisiae), worm (Caenorhabditis elegans), fly (Drosophila melanogaster), mouse (Mus musculus), Arabidopsis thaliana and human (Homo sapiens), and 747 data sets (276 co-expression networks, 232 physical interaction, 24 genetic interaction, 14 co-localization, 5 pathway, 176 predicted and 12 shared protein domain information). The network breakdown by organism is: yeast, 163; worm, 76; fly, 60; mouse, 129; Arabidopsis, 94; and human, 225. We currently support standard genes symbols; Ensembl, Entrez, UniProtKB, and RefSeq database identifiers; and unique gene synonyms. Since we use Ensembl as a primary identifier source, we do not recognize ambiguous gene names that map to multiple Ensembl genes within the same organism.
Data sources
Data sets are collected from publicly available databases, including co-expression data from Gene Expression Omnibus (GEO) (1); physical and genetic interaction data from BioGRID (2); predicted protein interaction data based on orthology from I2D (3); and pathway and molecular interaction data from Pathway Commons, which contains data from BioGRID (2), Memorial Sloan-Kettering Cancer Center, Human Protein Reference Database (4), HumanCyc (5), Systems Biology Center New York, IntAct (6), MINT (7), NCI-Nature Pathway Interaction Database (8) and Reactome (9). Individual data sets relevant to specific organisms are also collected, such as protein sub-cellular localization in yeast. Networks are produced from the data either directly (as in the case of protein or genetic interactions) or using an in-house analysis pipeline to convert profiles to functional association networks (e.g. mRNA expression data are converted to co-expression networks). This pipeline is described in detail in (10). In summary, co-expression networks are filtered to remove weak correlations, which we have shown decreases compute time without impacting prediction accuracy (10). A default set of networks has been selected for each organism; however, users can choose which networks to include in their analysis in the advanced options panel, found directly below the gene query text box on both the initial and results pages. Using this panel, users can select or deselect all networks from a given data source, or select or deselect individual networks, using a system of check boxes.
Uploading data sets
Users can upload their own data sets in tab-delimited text format to include in their analyses. The required format for uploaded data sets is described in ‘Upload Help’, which is beside the Upload button in the advanced options panel. These data sets are stored only during a user’s session, accessible only to the user who uploaded them and can be used seamlessly with the preloaded GeneMANIA data sets.
Network weighting methods
By default, the GeneMANIA prediction server uses one of two different adaptive network weighting methods. For longer gene lists, GeneMANIA uses the basic weighting method [called GeneMANIAEntry-1 in (10) and called ‘assigned based on query genes’ on the web site] and weights each network so that after the networks are combined, the query genes interact as much as possible with each other while interacting as little as possible with genes not in the list. GeneMANIA learns from longer gene lists, allowing a gene list-specific network weighting to be calculated. Shorter gene lists do not contain enough information for GeneMANIA to learn which networks mediate the underlying functional relationship among the genes. For short gene lists, GeneMANIA uses a similar principle to weight networks, but tries to reproduce Gene Ontology (GO) biological process co-annotation patterns rather than the gene list. This method is described in detail in (11). The user may choose other adaptive and non-adaptive weighting methods in the advanced options panel, found directly under the gene query text box. The two non-adaptive methods are the most conservative options and work well on small gene lists (10). These methods allow users to choose either to weight every individual network equally, or weight each class (e.g. co-expression and protein interaction) of network equally. Network weights can also be assigned based on how well they reproduce GO co-annotation patterns for that organism in the molecular function, biological process or cellular component hierarchies. Note that the annotation-based weighting may slightly inflate weights for networks on which current annotations are based or for networks that were derived based on co-annotation patterns of genes. The networks most affected by this inflation are the older, smaller scale protein and genetic interaction studies and networks classified as ‘predicted’. However, this inflation does not seem to have a large impact on weights and may be largely avoided by only using networks derived from high-throughput assays with the annotation-based schemes.
GENEMANIA OUTPUT
Determining the network weights
The constructed composite network is a weighted sum of individual data sources; each edge (link) in the composite network is weighted by the corresponding individual data source. The network weights are non-negative, sum to 100% and reflect the relevance of each data source for predicting membership in the query list. Given the composite network, we use label propagation (12) to score all non-query genes. These scores are used to rank the genes. The score assigned to each gene reflects how often paths that start at a given gene node end up in one of the query nodes and how long and heavily weighted those paths are. For more details on these algorithms, refer to (10). These scores are presented to the user in a table that allows interaction with the composite network display.
Effects of network selection
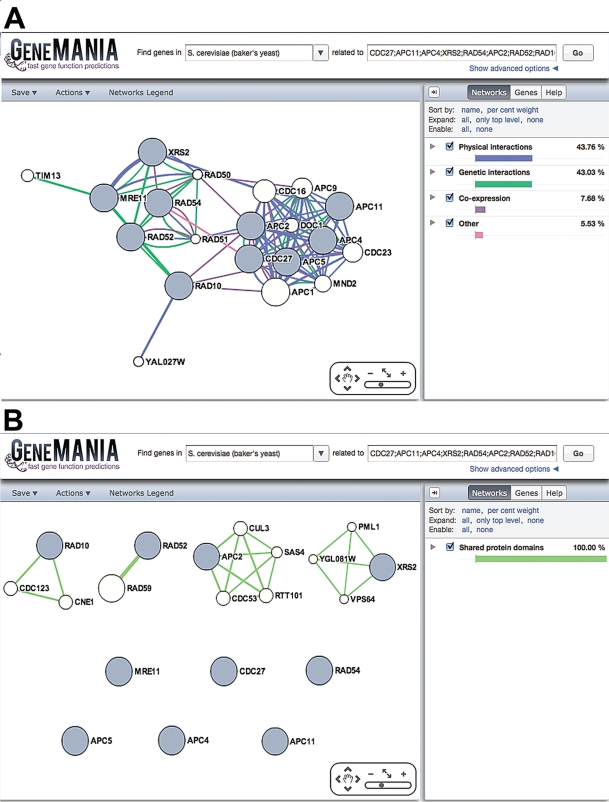
Choosing different parameters in GeneMANIA can change the results. For example, selecting different data sets to use in the analysis produces different networks. The yeast default query (cell cycle), using all default parameters is illustrated in Figure 2A. By only selecting shared protein domain networks, the network changes drastically (Figure 2B) where only genes that have the same protein domains are linked.
Figure 2.
Effects of data set selection on network topology. GeneMANIA showing the results of two yeast queries. (A) The yeast cell-cycle default query, using all default parameters. (B) The yeast cell-cycle default query, using default network weighting method. Only shared protein domain data sets are selected.
Effects of network weighting method selection
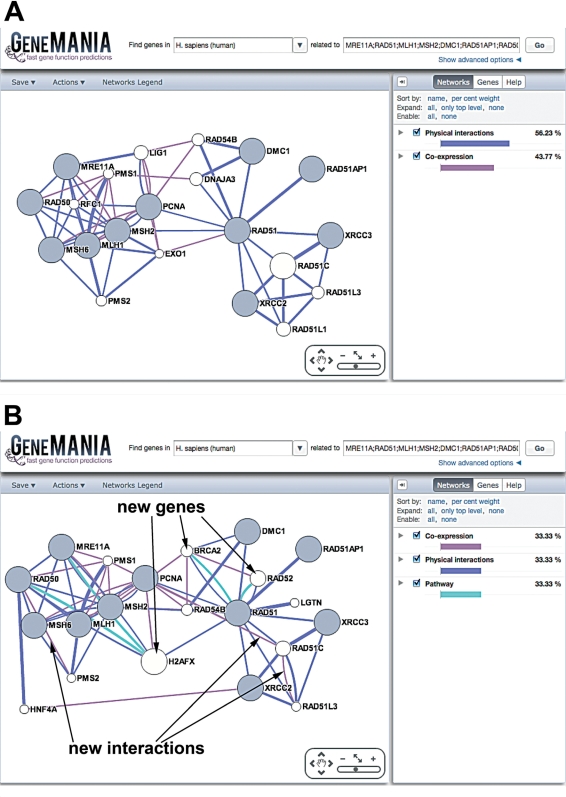
Different results can be produced from the same gene list and set of networks by changing the network weighting method, as illustrated in Figure 3 for a human DNA repair and replication and the default network selections. Figure 3A shows the results produced with the default network weighting method, the GeneMANIA algorithm (10). As none of the pathway data sets in GeneMANIA link members of the query gene list, the pathway category receives zero weight. In contrast, the network in Figure 3B was generated using the ‘Equal by data type’ network weighting method, which forces all selected network categories (i.e. data types) to be weighted equally, regardless of the number of networks selected in that category. The resulting networks are different, with different interactions and three different predicted genes that are linked to the query list by a pathway. These added levels of query customization enable users to tweak their results appropriately for their query.
Figure 3.
Effects of network weighting method selection on network topology. GeneMANIA showing the results of two human queries. (A) The human DNA repair and replication default query, using all default parameters. (B) The human DNA repair and replication default query, using the default data set selection but with the ‘Equal by data type’ network weighing method selected. Some genes and interactions found in this query that were not present in the default query (shown in A) are indicated by arrows.
Saving GeneMANIA results
GeneMANIA allows users to save the results of their analysis by clicking on ‘Save’ in the menu above the network. By selecting options in this menu, users can generate a report that includes any or all of the network image, the network weights, recommended genes and/or the query parameters. Users can also download the network weights and genes in the generated composite network as a tab-delimited text file.
ALGORITHM AND SOFTWARE VALIDATION
Tests of the predictive accuracy have been performed on all algorithms used by the GeneMANIA prediction server. The ‘assigned based on query genes’ weighting method boasts state-of-the-art performance on gene function prediction benchmarks in yeast (10) and mouse (13) for gene lists longer than 10 genes. The annotation-based weighting methods have state-of-the-art performance for smaller gene lists in a variety of model organisms (11). In Supplementary Figure S1, we compare the newer network weighting algorithms with the older ‘assigned based on query genes’ (called GM-2008 in Supplementary Figure S1) for each of the six organisms in GeneMANIA, using a range of data sets (Table 1), by assessing their ability to recover GO annotations in 10-fold cross-validation. Further details about assessing performance using shared GO terms are available in (11). The performance of the original GeneMANIA algorithm was used as a benchmark. The algorithms were tested on 12 282 GO terms across the six organisms. In many tests, we found the new methods perform as well, or better than, the original algorithm (Supplementary Figure S1). We have also extensively tested all GeneMANIA functionality using unit tests and functional testing across popular web browsers (Firefox 3, Safari and Internet Explorer 7) to ensure a high-quality user experience.
Table 1.
Number of networks per organism
| Network type | Organism |
|||||
|---|---|---|---|---|---|---|
| Arabidopsis thaliana | Worm | Fly | Human | Mouse | Yeast | |
| Co-expression | 56 | 10 | 40 | 69 | 46 | 55 |
| Physical interaction | 11 | 8 | 8 | 113 | 28 | 64 |
| Genetic interaction | 1 | 4 | 2 | 1 | 0 | 16 |
| Shared protein domains | 2 | 2 | 2 | 2 | 2 | 2 |
| Co-localization | 1 | 1 | 7 | 2 | 2 | 1 |
| Pathways | 0 | 0 | 0 | 5 | 0 | 0 |
| Predicted interaction | 23 | 50 | 1 | 33 | 48 | 21 |
| Other | 0 | 1 | 0 | 0 | 3 | 4 |
Co-expression and shared protein domain network links are weighted continuously from 0…1, physical and genetic interaction networks are binary. The ‘other’ category consists of organism-specific functional genomics networks, such as from the MouseFunc competition.
OTHER GENE FUNCTION PREDICTION PROGRAMS
Other gene function prediction web-based interfaces include N-Browse (14), the bioPIXIE system (15), MouseNet (16), STRING (17) and Functional Coupling (FunCoup) (18). The GeneMANIA prediction server offers a number of advantages in flexibility, data representation and predictive accuracy over these methods [see, e.g. (10)]. N-Browse boasts an elegant graphical user interface (GUI), with interactivity between the network display and node and edge information, and the networks involved in the analysis can be changed by the user; however, N-Browse functions as a Java web start, which is less convenient for the casual user. The bioPIXIE and MouseNET servers provide users with a fixed network (or networks) to query, built by incorporating multiple yeast and mouse genomic data sets. Similarly, STRING impressively supports 630 organisms, but gives users little choice about which functional association network data to use for their query. In contrast, the GeneMANIA prediction server generates results customized to the queried genes and the user-selected data sources so that the resulting network can be specifically tailored to the prediction task at hand. Although FunCoup has a similar functionality, it assigns weights using a naive Bayes framework that cannot detect redundancy among data sets. In addition to its ability to detect and compensate for data redundancy, GeneMANIA’s prediction server also has an advantage due to the state-of-the-art predictive accuracy of its label propagation algorithm (10,13). Direct comparison of predictive accuracy of GeneMANIA with other web servers is difficult, as every server collects different data sets and processes them differently. However, GeneMANIA is faster and achieves higher area under the receiver-operator characteristic (ROC) curve than bioPIXIE in predicting GO annotation using only 5 (of GeneMANIA’s 163) yeast networks (10) and performs very well against a suite of gene function prediction methods (most of which are too slow to be deployed in a web server) in a carefully controlled function prediction challenge in mouse (13).
CONCLUSIONS
We have developed GeneMANIA for gene function predictions, consisting of a highly adaptive algorithm, easily extendable database and interactive, intuitive interface. In addition to the introduction presented here, we have an extensive user manual that is accessible by clicking ‘Help’ on the initial query page, or in the bottom left row in the network browser. We are currently working with OpenHelix to develop an online tutorial for GeneMANIA. Our functional prediction algorithm is organism independent and remains an area of active research in our laboratories. We are currently investigating modifications to make it faster and more accurate which we expect to have integrated by the end of the year. We currently support six organisms, but over time we will be adding additional model organisms and new large-scale data sets as they become available. GeneMANIA is an open source project. Please contact the corresponding authors for access to the source code.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Genome Canada through the Ontario Genomics Institute (2007-OGI-TD-05), with additional support by a Natural Science and Engineering Research Council of Canada operating grant (to Q.M.); Undergraduate Student Research Award award (to D.W.F.). Funding for open access charge: Genome Canada through the Ontario Genomics Institute (2007-OGI-TD-05).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Ruth Isserlin and the Emili Lab for help collecting S. cerevisiae data sets, and Nicholas Provart for contributing A. thaliana data sets compiled by his lab.
REFERENCES
- 1.Barrett T., Troup D.B., Wilhite S.E., Ledoux P., Rudnev D., Evangelista C., Kim I.F., Soboleva A., Tomashevsky M., Marshall K.A., et al. NCBI GEO: archive for high-throughput functional genomic data. Nucleic Acids Res. 2009;37:D885–D890. doi: 10.1093/nar/gkn764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Breitkreutz B.J., Stark C., Reguly T., Boucher L., Breitkreutz A., Livstone M., Oughtred R., Lackner D.H., Bähler J., Wood V., et al. The BioGRID Interaction Database: 2008 update. Nucleic Acids Res. 2008;36:D637–D640. doi: 10.1093/nar/gkm1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brown K.R., Jurisica I. Online predicted human interaction database. Bioinformatics. 2005;21:2076–2082. doi: 10.1093/bioinformatics/bti273. [DOI] [PubMed] [Google Scholar]
- 4.Keshava Prasad T.S., Goel R., Kandasamy K., Keerthikumar S., Kumar S., Mathivanan S., Telikicherla D., Raju R., Shafreen B., Venugopal A., et al. Human Protein Reference Database–2009 update. Nucleic Acids Res. 2009;37:D767–D772. doi: 10.1093/nar/gkn892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Romero P., Wagg J., Green M.L., Kaiser D., Krummenacker M., Karp P.D. Computational prediction of human metabolic pathways from the complete human genome. Genome Biol. 2005;6:R2. doi: 10.1186/gb-2004-6-1-r2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Aranda B., Achuthan P., Alam-Faruque Y., Armean I., Bridge A., Derow C., Feuermann M., Ghanbarian A.T., Kerrien S., Khadake J., et al. The IntAct molecular interaction database in 2010. Nucleic Acids Res. 2010;38:D525–D531. doi: 10.1093/nar/gkp878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ceol A., Chatr Arayamontri A., Licata L., Peluso D., Briganti L., Perfetto L., Castagnoli L., Cesareni G. MINT, the molecular interaction database: 2009 update. Nucleic Acids Res. 2010;38:D532–D539. doi: 10.1093/nar/gkp983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schaefer C.F., Anthony K., Krupa S., Buchoff J., Day M., Hannay T., Buetow K.H. PID: the Pathway Interaction Database. Nucleic Acids Res. 2009;37:D674–D679. doi: 10.1093/nar/gkn653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vastrik I., D'Eustachio P., Schmidt E., Gopinath G., Croft D., de Bono B., Gillespie M., Jassal B., Lewis S., Matthews L., et al. Reactome: a knowledge base of biologic pathways and processes. Genome Biol. 2007;8:R39. doi: 10.1186/gb-2007-8-3-r39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mostafavi S., Ray D., Warde-Farley D., Grouios C., Morris Q.D. GeneMANIA: a real-time multiple association network integration algorithm for predicitng gene function. Genome Biol. 2008;9:S4. doi: 10.1186/gb-2008-9-s1-s4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mostafavi S., Morris Q. Fast integration of heterogeneous data sources for predicting gene function with limited annotation. Bioinformatics. 2010 doi: 10.1093/bioinformatics/btq262. [27 May 2010, Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhou D., Bousquet O., Lal T., Weston J., Scholkopf B. Learning with local and global consistency. In: Thrun S., Saul K., Scholkopf B., editors. Advances in Neural Information Processing Systems 16. Cambridge: MIT Press; 2003. pp. 321–328. [Google Scholar]
- 13.Pena-Castillo L., Tasan M., Myers C.L., Lee H., Joshi T., Zhang C., Guan Y., Leone M., Pagnani A., Kim W.K., et al. A critical assessment of Mus musculus gene function prediction using integrated genomic evidence. Genome Biol. 2008;9:S2. doi: 10.1186/gb-2008-9-s1-s2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kao H.-L., Gunsalus K.C. Browsing multidimensional molecular networks with the generic network browser (N-Browse) Curr. Protoc. Bioinform. 2008;23 doi: 10.1002/0471250953.bi0911s23. 9.11.1-9.11.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Myers C.L., Robson D., Wible A., Hibbs M.A., Chiriac C., Theesfeld C.L., Dolinski K., Troyanskaya O.G. Discovery of biological networks from diverse functional genomic data. Genome Biol. 2005;6:R114. doi: 10.1186/gb-2005-6-13-r114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guan Y., Myers C.L., Lu R., Lemischka I.R., Bult C.J., Troyanskaya O.G. A genomewide functional network for the laboratory mouse. PLoS Comput. Biol. 2008;4:1–15. doi: 10.1371/journal.pcbi.1000165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jensen L.J., Kuhn M., Stark M., Chaffron S., Creevey C., Muller J., Doerks T., Julien P., Roth A., Simonovic M., et al. STRING 8 – a global view on proteins and their functional interactions in 630 organisms. Nucleic Acids Res. 2009;37:D412–D416. doi: 10.1093/nar/gkn760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Alexeyenko A., Sonhammer E.L.L. Global networks of functional coupling in eukaryotes from comprehensive data integration. Genome Res. 2009;19:1107. doi: 10.1101/gr.087528.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Toufighi K., Brady S.M., Austin R., Ly E., Provart N.J. The Botany Array Resource: e-Northerns, Expression Angling, and promoter analyses. Plant J. 2005;43:153–163. doi: 10.1111/j.1365-313X.2005.02437.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.