Abstract
Gene set enrichment analysis (GSEA) is a widely used technique in transcriptomic data analysis that uses a database of predefined gene sets to rank lists of genes from microarray studies to identify significant and coordinated changes in gene expression data. While GSEA has been playing a significant role in understanding transcriptomic data, no similar tools are currently available for understanding metabolomic data. Here, we introduce a web-based server, called Metabolite Set Enrichment Analysis (MSEA), to help researchers identify and interpret patterns of human or mammalian metabolite concentration changes in a biologically meaningful context. Key to the development of MSEA has been the creation of a library of ∼1000 predefined metabolite sets covering various metabolic pathways, disease states, biofluids, and tissue locations. MSEA also supports user-defined or custom metabolite sets for more specialized analysis. MSEA offers three different enrichment analyses for metabolomic studies including overrepresentation analysis (ORA), single sample profiling (SSP) and quantitative enrichment analysis (QEA). ORA requires only a list of compound names, while SSP and QEA require both compound names and compound concentrations. MSEA generates easily understood graphs or tables embedded with hyperlinks to relevant pathway images and disease descriptors. For non-mammalian or more specialized metabolomic studies, MSEA allows users to provide their own metabolite sets for enrichment analysis. The MSEA server also supports conversion between metabolite common names, synonyms, and major database identifiers. MSEA has the potential to help users identify obvious as well as ‘subtle but coordinated’ changes among a group of related metabolites that may go undetected with conventional approaches. MSEA is freely available at http://www.msea.ca.
INTRODUCTION
Metabolomics is a field of omics science concerned with the comprehensive characterization of small molecule metabolites found in cells, tissues, biofluids, and organisms. It uses a combination of NMR spectroscopy, mass spectrometry, and/or liquid/gas chromatography to specifically identify metabolites or generate metabolic spectral profiles. Because metabolomics is concerned with looking at the small molecule products of gene, protein, and environmental interactions, it provides complementary information to what is normally obtained via genomics, transcriptomics, and proteomics. As a consequence, metabolomics is playing an increasingly important role in both systems biology and synthetic biology (1,2). It is also finding wide applications in diagnostic biomarker discovery, toxicological testing, food and beverage analysis, plant and animal phenotyping as well as drug discovery and development (1–4).
There are two routes to conducting a metabolomics experiment. One is called quantitative (or targeted) metabolomics and the other is called chemometric (or untargeted) metabolomics. In chemometric metabolomics, spectral patterns from two or more large sample sets are processed chemometrically and significant peak differences are identified. The limited number of compounds contributing to these differences is then (ideally) identified. In quantitative metabolomics, large numbers of compounds are first identified and quantified before the data are further processed. In this regard, quantitative metabolomics is more similar to a standard proteomics or transcriptomics experiment. As with any ‘omics’ experiment, a typical quantitative metabolomic study consists of three stages: data collection, data analysis, and data interpretation. In the data collection stage, sample spectra are first acquired using various analytical platforms (NMR, GC-MS, LC-MS, HPLC). These spectra are then processed by different software tools to facilitate compound identification and quantification, thereby generating metabolite lists. In the second (data analysis) stage, various statistical methods are applied to identify those metabolites that are changed significantly under the given study conditions. Popular methods include t-tests, principal component analysis (PCA), partial least square (PLS) discriminant analysis, as well as a variety of other methods. Compounds are first ranked using appropriate importance measures such as P-values, loading scores or variable importance in projection scores. A cut-off threshold is then applied to select the top n metabolites from the ranked list. In the final stage (data interpretation), the list of significant metabolites is examined, often manually, to see if any biological interesting patterns can be identified. For example, if compounds involved in a certain metabolic pathway appear to be more frequently observed than would be anticipated by random chance, then it may be reasonable to assume that this pathway is biologically or metabolically important.
There are several potential problems associated with this kind of analytical protocol, particularly with the second and third stages. In the second stage, the threshold used for selecting important metabolites is usually chosen arbitrarily. Many moderate but meaningful changes may be missed if an inappropriate threshold is chosen. Indeed, compounds that are critical components of a particular biological process may be left out and the resulting patterns could become indiscernible in the third stage. Choosing a different threshold value may, therefore, lead to a different biological conclusion. In addition, most statistical methods used in compound selection assume that metabolites are being sampled independently, which is certainly not true given the intricate correlations and connections seen among metabolites in metabolic networks. In the third stage, despite obvious differences in importance measures, all selected compounds are treated equally, as only their occurrences (and not their concentrations) are considered in the analysis. This loss of information can obviously reduce the accuracy of any subsequent interpretations. In addition, since the identification of metabolically meaningful patterns is usually performed manually, this process can be very time consuming. Likewise, the final interpretation is often subject to the background knowledge or biases of individual researchers.
These data analysis/interpretation issues are not unique to metabolomics. In fact, they have long been the subject of intense debate among researchers involved in gene expression data analysis (5–8). This debate has inspired the development of a new approach for gene expression analysis, generally referred to as gene set enrichment analysis (GSEA). The key idea behind GSEA is to directly investigate the enrichment of pre-defined groups of functionally related genes (or gene sets) instead of individual genes. This group-based approach does not require pre-selection of genes with an arbitrary threshold. Instead, functionally related genes are evaluated together as gene sets, allowing additional biological information to be incorporated into the analysis process. The GSEA approach has proven to be remarkably successful in deriving new information from genome-wide expression studies, having been cited 1000′s of times since its initial description in 2005 (9). Its success has also inspired many extensions, improvements and variations (8–16).
However, to our knowledge, no tools similar to GSEA have been developed to support this group-based approach for metabolomic data analysis. This is likely because both enrichment analysis and quantitative metabolomics are relatively new techniques. However, it is also likely due to the fact that in order to use this approach, one needs an extensive and biologically meaningful metabolite set library. Such a library is very laborious and time consuming to create. To address this issue, we collected, both through text-mining and manual curation, a large body of mammalian (primarily human) metabolite and metabolic pathway information from the literature and various public databases. Using this library of metabolites and disease/pathway/tissue associations, we have implemented a web-based application, named MSEA, to support group-based enrichment analysis for human and/or mammalian metabolomic studies. The main features of MSEA include the following.
A collection of five metabolite set libraries containing ∼1000 biologically meaningful groups of metabolites.
Three enrichment analysis methods – overrepresentation analysis (ORA), single sample profiling (SPP), and quantitative enrichment analysis (QEA), to support common data forms generated in metabolomic studies;
Support for enrichment analysis with discrete and continuous phenotypes.
Support for enrichment analysis using customized (non-mammalian) metabolite sets.
Support for conversions between metabolite common names, synonyms, and identifiers (ID) of nine major metabolomic databases.
Comprehensive analysis report generation.
In other words, with MSEA and its accompanying databases it is possible to take a list of altered metabolites from a biofluid or tissue sample and use it to suggest a biological pathway or disease condition that can be further investigated. The MSEA server and all of its accompanying databases are freely available at http://www.msea.ca.
METHODS
Creation of metabolite set libraries
A group of metabolites are considered to constitute a metabolite set if they are known to be: (1) involved in the same biological processes (i.e., metabolic pathways, signaling pathways); (2) changed significantly under the same pathological conditions (i.e., various metabolic diseases); and (3) present in the same locations such as organs, tissues, or cellular organelles. These data were collected through manual curation from books and journals as well as through text mining of public databases. The resulting metabolite sets were manually validated/edited and then further organized into three categories: pathway associated, disease associated, and location based. MSEA’s pathway-associated metabolite library contains 84 entries based on the 84 human metabolic pathways found in the Small Molecular Pathway Database (SMPDB) (17). MSEA’s disease-associated metabolite sets were mainly collected from the literature. Metabolites associated with different diseases were manually identified, merged and subsequently refined by reading the original publications listed in the Human Metabolome Database (HMDB) (18), the Metabolic Information Center (MIC), and SMPDB. Using these resources, a total of 851 physiologically informative metabolite sets were created. These disease-associated metabolite sets were further divided into three subcategories based on the biofluids in which they were measured: 398 metabolite sets in blood; 335 in urine; and 118 in cerebral–spinal fluid (CSF). MSEA’s location-based library contains 57 metabolite sets based on the ‘Cellular Location’ and ‘Tissue Location’ listed in the HMDB. A summary of these metabolite set libraries is shown in Table 1.
Table 1.
Overview of MSEA’s metabolite set libraries
| Category | Total number | Sources | Web links |
|---|---|---|---|
| Pathway based | 84 | SMPDB | http://www.smpdb.ca |
| Disease—associated | 851 | ||
| Blooda | 398 | HMDB | http://www.hmdb.ca/disease_browse |
| Urinea | 335 | MIC | http://www.metagene.de |
| CSFa | 118 | PubMed | http://www.ncbi.nlm.nih.gov/pubmed |
| Location based | 57 | HMDB | http://www.hmdb.ca/ |
aMetabolite sets were collected from multiple sources including HMDB, MIC, PubMed and SMPDB.
Creation of a metabolite dictionary and concentration database
In order for the MSEA server to accept a range of metabolite names, synonyms or ID as input, it was also necessary to develop a local metabolite dictionary that could be used to perform facile name conversion or ‘normalization’. Information contained in the HMDB was used to extract common names, synonyms, as well as ID used in nine major metabolomic databases [HMDB, PubChem (19), ChEBI (20), KEGG (21), BiGG (22), METLIN (23), BioCyc (24), Reactome (25), and Wikipedia]. Examples of MSEA’s supported IDs are listed in Table 2. In order for MSEA to perform single sample profiling (SSP) analysis, it was also critical to obtain reference concentrations for as many metabolites as possible. These concentration data were collected primarily from the HMDB with additional values being added through manual curation. MSEA’s reference concentrations are organized based on the biofluids in which they were measured. Concentrations are presented in the form of mean (minimum – maximum). For concentrations reported as mean and standard deviation (SD), their 95% confidence intervals (mean ± 2 SD) were used to define the concentration ranges. One compound may have multiple concentration values as reported from different studies.
Table 2.
Overview of compound labels currently supported by MSEA
| Label type | Examples |
|---|---|
| Common Name | Adenosine, acetic acid, adenine, creatine |
| HMDB | HMDB00050, HMDB00042, HMDB00034, HMDB0006 |
| PubChem | 60961, 176, 190, 586 |
| ChEBI | 16335, 15366, 16708, 16919 |
| KEGG | C00212, C00033, C00147, C00300 |
| BiGG | 34273, 33590, 34039, 34543 |
| METLIN | 86, 3206, 85, 7 |
| BioCyc | ADENOSINE, ACET, ADENINE, CREATINE |
| Reactome | 114933, 114747, 114936, 114818 |
| Wikipedia | Adenosine, acetic acid, adenine, creatine |
Implementation of enrichment analysis programs
Over the past 5 years, many different algorithms have been developed for group-based enrichment analysis, including GSEA (9), GSEA-P (26), PAGE (27), globaltest (11), SAFE (12), SAM-GS (13) and GSA (14). Based on a thorough review of the literature, we decided to adopt the globaltest algorithm as the backend for MSEA. There were three main reasons: (1) recent publications have indicated that globaltest exhibited similar or superior performance when tested against several other algorithms (28–30); (2) globaltest is very flexible and supports binary, multiclass, and continuous phenotype labels; and (3) globaltest is computationally efficient as the P-values can be calculated based on the Q-stat’s asymptotic distribution, which appears to work well with both large and small sample sizes. The globaltest algorithm was originally designed for testing associations between gene sets and clinical outcomes (11). It uses a generalized linear model to compute a ‘Q-stat’ for each gene set. For a group of m genes, the Q-stat is calculated as the average of the Q values (Q1…Qm) calculated for the m single genes, where Qi is the average of the squared covariance between the gene expression pattern and the clinical outcome. Conventional ORA was implemented based on a cumulative hypergeometric distribution. Since many metabolite sets are tested simultaneously, we also implemented methods to adjust for the multiple testing problems that occur during enrichment analysis. In addition to the original P-values, MSEA also reports Bonferroni corrected P-values and a false discovery rate (FDR) according to Benjamini and Hochberg (31).
Web server characteristics
MSEA’s web interface was implemented using the JSF or Java Server Faces (http://java.sun.com/javaee/javaserverfaces) framework. The enrichment analysis algorithms were implemented in the R (version 2.10.0) programming language (http://www.r-project.org/). The communication between R and Java was established through the Rserve TCP/IP server (http://www.rforge.net/Rserve/). The web application is hosted on GlassFish (version 3) using a Linux operating system (Fedora Core 10). MSEA’s host server is equipped with two Intel Quad Core 2 processors (3.0 GHz each) and 8 GB of physical memory. The web application is platform independent and has been tested successfully on Internet Explorer 8.0, Mozilla Firefox 3.0, and Safari 4.0.
PROGRAM DESCRIPTION
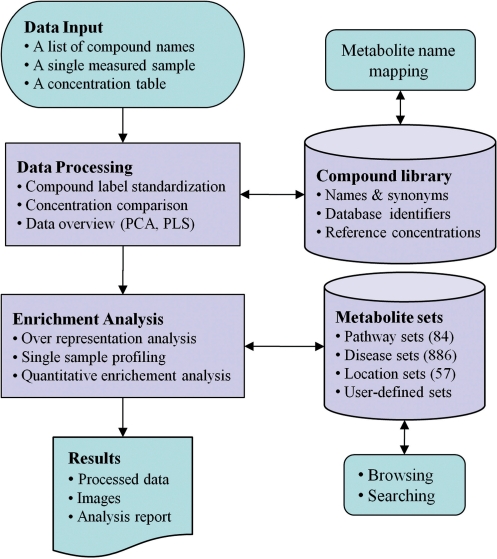
MSEA’s workflow is illustrated in Figure 1. Briefly, MSEA can be described in four steps – data input, data processing, data analysis, and data download. In addition to its analysis utilities, users can directly download, browse or search MSEA’s metabolite set libraries, or perform compound name and ID conversions. The details of each step are discussed below.
Figure 1.
MSEA workflow. MSEA consists of four steps—data input, data processing, data analysis, and data download. Different analysis procedures are performed for different input types. MSEA allows users to directly browse and search its metabolite set libraries as well as to perform metabolite name mapping between different names and database ID.
Step 1. Data input
MSEA accepts data in three different formats: (i) a list of compound names entered in a single-column format; (ii) a list of compound concentrations entered as two-column data with the first column corresponding to the compound names/labels and the second corresponding to the concentration values; or (iii) a concentration table containing metabolite concentration data from multiple samples. The table must contain comma-separated values (.csv) with rows for samples and columns for metabolites. The second column of the table is reserved for phenotype labels (binary, multiclass, or continuous). Examples of these input formats are provided on the MSEA homepage.
Step 2. Data processing
In this step, both the compound labels and the concentration values are examined for their suitability for downstream analysis. It is critical that the compound labels be recognized by the program in order to be compared with MSEA’s collection of compound names in metabolite sets. Therefore, a consistency check is done with the input names or IDs against the names and IDs stored in MSEA’s metabolite dictionary. Any nomenclature inconsistency is flagged and displayed to users for manual inspection and correction. For SSP, the concentrations must be provided in a standard concentration unit (µmol for blood and CSF and µmol mmol−1_creatinine for urine) in order for the input data to be properly compared with MSEA’s reference concentrations database. For QEA, the concentration values can be normalized and negative/missing values are allowed. Two widely used chemometric methods—PCA and PLS analysis—are available in MSEA to allow for data visualization, pattern identification, and outlier detection. Note that MSEA does not perform data normalization. Users are advised to visit MSEA’s companion web site MetaboAnalyst (32) to access a variety of data processing and normalization options.
Step 3. Enrichment analysis
Depending on the type of user input, MSEA offers three kinds of enrichment analysis: ORA; SSP, and QEA. These analysis modules are described in more detail below.
ORA
ORA is used to evaluate whether a particular set of metabolites is represented more than expected by chance within a given compound list. ORA is performed when the user provides only a list of compound names. Such a list can be obtained using standard feature selection methods that statistically rank all the compounds and select those scoring above a certain threshold. ORA is also very useful for analyzing a group of compounds exhibiting similar concentration changes or patterns. Such a list can be obtained from standard clustering analysis. Many commonly used feature selection and feature clustering methods are available from our companion web application MetaboAnalyst (32). The P-value from ORA indicates the probability of seeing at least a particular number of metabolites from a certain metabolite set in a given compound list. The Bonferroni corrected P-value and FDR are also presented to account for problems associated with multiple comparisons. Users can click the ‘View’ link in the Details column of any of MSEA’s metabolite sets to see all its constituent metabolites with matched ones highlighted in red, as well as pathway images (when available).
SSP
For common human biofluids such as blood, urine, or CSF, normal concentration ranges are known for many metabolites. In clinical metabolomic studies, it is often desirable to know whether certain metabolite concentrations in a given sample are significantly higher or lower than their normal ranges. MSEA’s SSP module is designed to provide this kind of analysis. In particular, SSP is performed when the user provides a two-column list of both compounds and concentrations. When called, the SSP module will compare the measured concentration values of each compound to its recorded normal reference ranges of the corresponding biofluid (Figure 2A). By default, only compounds with concentrations above or below all the reported normal ranges will be selected for further investigation. Users can manually select or deselect compounds to override this default selection by inspecting the concentration comparison plots generated by this module (Figure 2B).
Figure 2.
Enrichment analysis and visualization. Results from MSEA’s enrichment analysis are presented both in tables as well as through graphical summaries. (A) The comparison between the measured concentrations and reference concentrations using the SSP module. The top part of (B) shows a graphical summary of the concentration comparison for a single compound when users click an image icon in Figure 2A. The bottom part of Figure 2B shows all the corresponding publications that reported these concentrations. (C) The results generated by the QEA module. The top part of (D) is a metabolite-set plot indicating the influence of an individual compound on each of the selected metabolite sets. The bottom part of Figure 2D shows all its constituent metabolites with matched ones highlighted in red.
QEA
QEA is performed when the user uploads a concentration table containing metabolite concentration data from multiple samples. QEA is based on the globaltest algorithm to perform enrichment analysis directly from raw concentration data and does not require a list of significantly changed compounds. With QEA, enriched metabolite sets can be identified when only a few compounds are significantly changed or when many compounds are only slightly (but consistently) changed. The QEA algorithm uses a generalized linear model to estimate a ‘Q-stat’ for each metabolite set. The Q-stat describes the correlation between compound concentration profiles, X, and phenotype labels, Y. In addition to the Q-stat values, the QEA module also provide P-values, Bonferroni-corrected P-values, and estimates of FDR. Figure 2C shows a screenshot of the output table from a typical QAE. Users can click the image icon of any matched metabolite set to view a detailed graphical summary of the contributions of individual metabolites (Figure 2D).
Step 4. Data download
When users finish an enrichment analysis, a comprehensive report is generated with detailed descriptions of each step performed, embedded with graphical and tabular results. The processed data, images, R scripts, as well as the R command history are also available for download. Users familiar with R can easily reproduce the results on their local machine after installing the R packages and the corresponding metabolite set libraries (available on the Resources Download page).
Other features
The MSEA web server also offers a number of other features to facilitate metabolomic data analysis, including (1) a compound name and ID mapping tool; (2) a browser for metabolite sets; and (3) a facility for custom metabolite set uploads. Given the fact that no consensus exists in labeling compounds in current metabolomic studies, we implemented a utility in MSEA to convert between common compound names, synonyms and the ID codes used in nine major metabolite databases (see Table 2 for details). This converter can also deal with spelling errors using an approximate text matching algorithm. In addition to this name/ID converter, MSEA also provides a browser to view MSEA’s collection of metabolite set libraries. These libraries can provide a valuable source of information to investigate the biological implications of any metabolite sets identified after enrichment analysis. The browser implemented in the MSEA web server allows users to easily scan and search its metabolite set libraries. Each entry contains the metabolite set name, its constituent compounds, and links to original references. Given the incompleteness of MSEA’s metabolite-set libraries, researchers may want to perform enrichment analysis using customized or self-defined metabolite sets other than the ones provided by the server. MSEA supports this option by allowing users to upload their own metabolite set library. The library file should be in a simple.csv file with the first column for metabolite set names and the second for compound members.
Limitations
Unlike genomics or transcriptomics, metabolomics has not yet achieved total metabolite coverage. Whereas Next-Gen DNA sequencers and modern microarrays routinely cover entire genomes, most metabolomic technologies only offer 5–10% coverage of a sample’s metabolome (1–2). This makes many metabolomic studies intrinsically biased. Since most of the metabolite sets in MSEA’s libraries are also derived from experimental studies, they tend to suffer the same sampling bias. Fortunately, these biases tend to cancel each other out, as essentially the same metabolite population (the fraction of the metabolome that are ‘detectable’ by current analytical technologies) is probed to generate both metabolite sets and user data. Nevertheless, users should always take note of their experimental conditions or technological limitations when interpreting the results from enrichment analysis.
Another key limitation to MSEA is its bias to human and/or mammalian metabolomics. This is a limitation that we are working to overcome through the addition of other metabolite sets from plants and microbes. However, until these databases and data sets can be completed (likely in two years time) we would encourage researchers who are engaged in metabolomic studies of non-mammalian species to create their own customized metabolite sets for enrichment analysis and to contribute these sets to the MSEA server for public use.
CONCLUSIONS
Over the past few years, a number of software tools have been developed to address the bioinformatic needs of metabolomics. However, most of these programs were designed for spectral data processing and compound identification. More recently, several freely available software tools for the statistical analysis of metabolomic data have started to appear, such as MetaboAnalyst (32) and MeltDB (33). As yet, no publicly available tools have been made available to assist in the functional or biological interpretation of metabolomic data. To address this issue, we have developed a web server, named MSEA, designed to help researchers identify and interpret patterns of metabolite concentration changes in a biologically meaningful context. MSEA performs three kinds of enrichment analysis including ORA, SSP, and QEA. When only a list of compounds is available ORA is performed. When both compound names and concentrations are available the SSP module is called. When concentration data are available from multiple samples, MSEA performs QEA. The enrichment analyses performed by MSEA are based on five carefully compiled metabolite libraries consisting of ∼1000 entries. In addition to its enrichment analysis capabilities, MSEA allows custom metabolite sets to be uploaded for more specialized (non-mammalian) studies. MSEA also supports conversion between metabolite common names, synonyms and major database ID. We believe that, over time, the MSEA approach will become more powerful as analytical technologies for metabolomics continue to improve their metabolite coverage and as the metabolomics community develops improved standards and ontologies (34). In the long run, we would like to turn the MSEA server into a resource for metabolomic annotation, visualization, and integrated discovery much as the DAVID server (35) has become just such a resource for microarray data analysis.
FUNDING
Alberta Ingenuity Fund, the Alberta Advanced Education and Technology; the Canadian Institutes for Health Research; Genome Alberta, a division of Genome Canada. Funding for open access charge: Canadian Institutes for Health Research
Conflict of interest statement. None declared.
REFERENCES
- 1.Wishart DS. Quantitative metabolomics using NMR. Trends Anal. Chem. 2008;27:228–237. [Google Scholar]
- 2.Hollywood K, Brison DR, Goodacre R. Metabolomics: current technologies and future trends. Proteomics. 2006;6:4716–4723. doi: 10.1002/pmic.200600106. [DOI] [PubMed] [Google Scholar]
- 3.Ewald JC, Heux S, Zamboni N. High-throughput quantitative metabolomics: workflow for cultivation, quenching, and analysis of yeast in a multiwell format. Anal. Chem. 2009;81:3623–3629. doi: 10.1021/ac900002u. [DOI] [PubMed] [Google Scholar]
- 4.Gieger C, Geistlinger L, Altmaier E, Hrabe de Angelis M, Kronenberg F, Meitinger T, Mewes HW, Wichmann HE, Weinberger KM, Adamski J, et al. Genetics meets metabolomics: a genome-wide association study of metabolite profiles in human serum. PLoS Genet. 2008;4:e1000282. doi: 10.1371/journal.pgen.1000282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Khatri P, Draghici S. Ontological analysis of gene expression data: current tools, limitations, and open problems. Bioinformatics. 2005;21:3587–3595. doi: 10.1093/bioinformatics/bti565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pan KH, Lih CJ, Cohen SN. Effects of threshold choice on biological conclusions reached during analysis of gene expression by DNA microarrays. Proc. Natl Acad. Sci. USA. 2005;102:8961–8965. doi: 10.1073/pnas.0502674102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nam D, Kim SY. Gene-set approach for expression pattern analysis. Brief. Bioinform. 2008;9:189–197. doi: 10.1093/bib/bbn001. [DOI] [PubMed] [Google Scholar]
- 8.Mootha VK, Lindgren CM, Eriksson KF, Subramanian A, Sihag S, Lehar J, Puigserver P, Carlsson E, Ridderstrale M, Laurila E, et al. PGC-1 alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat. Genet. 2003;34:267–273. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- 9.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl Acad. Sci. USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee HK, Braynen W, Keshav K, Pavlidis P. ErmineJ: tool for functional analysis of gene expression data sets. BMC Bioinformatics. 2005;6:269. doi: 10.1186/1471-2105-6-269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Goeman JJ, van de Geer SA, de Kort F, van Houwelingen HC. A global test for groups of genes: testing association with a clinical outcome. Bioinformatics. 2004;20:93–99. doi: 10.1093/bioinformatics/btg382. [DOI] [PubMed] [Google Scholar]
- 12.Barry WT, Nobel AB, Wright FA. Significance analysis of functional categories in gene expression studies: a structured permutation approach. Bioinformatics. 2005;21:1943–1949. doi: 10.1093/bioinformatics/bti260. [DOI] [PubMed] [Google Scholar]
- 13.Dinu I, Potter JD, Mueller T, Liu Q, Adewale AJ, Jhangri GS, Einecke G, Famulski KS, Halloran P, Yasui Y. Improving gene set analysis of microarray data by SAM-GS. BMC Bioinformatics. 2007;8:242. doi: 10.1186/1471-2105-8-242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Efron B, Tibshirani R. On testing the significance of sets of genes. Ann. Appl. Stat. 2007;1 [Google Scholar]
- 15.Backes C, Keller A, Kuentzer J, Kneissl B, Comtesse N, Elnakady YA, Muller R, Meese E, Lenhof HP. GeneTrail–advanced gene set enrichment analysis. Nucleic Acids Res. 2007;35:W186–W192. doi: 10.1093/nar/gkm323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zheng Q, Wang XJ. GOEAST: a web-based software toolkit for Gene Ontology enrichment analysis. Nucleic Acids Res. 2008;36:W358–W363. doi: 10.1093/nar/gkn276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Frolkis A, Knox C, Lim E, Jewison T, Law V, Hau DD, Liu P, Gautam B, Ly S, Guo AC, et al. SMPDB: The Small Molecule Pathway Database. Nucleic Acids Res. 2010;38:D480–D487. doi: 10.1093/nar/gkp1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wishart DS, Tzur D, Knox C, Eisner R, Guo AC, Young N, Cheng D, Jewell K, Arndt D, Sawhney S, et al. HMDB: the human metabolome database. Nucleic Acids Res. 2007;35:D521–D526. doi: 10.1093/nar/gkl923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Austin CP, Brady LS, Insel TR, Collins FS. NIH Molecular Libraries Initiative. Science. 2004;306:1138–1139. doi: 10.1126/science.1105511. [DOI] [PubMed] [Google Scholar]
- 20.Degtyarenko K, de Matos P, Ennis M, Hastings J, Zbinden M, McNaught A, Alcantara R, Darsow M, Guedj M, Ashburner M. ChEBI: a database and ontology for chemical entities of biological interest. Nucleic Acids Res. 2008;36:D344–D350. doi: 10.1093/nar/gkm791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kanehisa M, Goto S, Kawashima S, Okuno Y, Hattori M. The KEGG resource for deciphering the genome. Nucleic Acids Res. 2004;32:D277–280. doi: 10.1093/nar/gkh063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Feist AM, Herrgard MJ, Thiele I, Reed JL, Palsson BO. Reconstruction of biochemical networks in microorganisms. Nat. Rev. Microbiol. 2009;7:129–143. doi: 10.1038/nrmicro1949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Smith CA, O'Maille G, Want EJ, Qin C, Trauger SA, Brandon TR, Custodio DE, Abagyan R, Siuzdak G. METLIN – A metabolite mass spectral database. Ther. Drug Monit. 2005;27:747–751. doi: 10.1097/01.ftd.0000179845.53213.39. [DOI] [PubMed] [Google Scholar]
- 24.Karp PD, Ouzounis CA, Moore-Kochlacs C, Goldovsky L, Kaipa P, Ahren D, Tsoka S, Darzentas N, Kunin V, Lopez-Bigas N. Expansion of the BioCyc collection of pathway/genome databases to 160 genomes. Nucleic Acids Res. 2005;33:6083–6089. doi: 10.1093/nar/gki892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Matthews L, Gopinath G, Gillespie M, Caudy M, Croft D, de Bono B, Garapati P, Hemish J, Hermjakob H, Jassal B, et al. Reactome knowledgebase of human biological pathways and processes. Nucleic Acids Res. 2009;37:D619–D622. doi: 10.1093/nar/gkn863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Subramanian A, Kuehn H, Gould J, Tamayo P, Mesirov JP. GSEA-P: a desktop application for Gene Set Enrichment Analysis. Bioinformatics. 2007;23:3251–3253. doi: 10.1093/bioinformatics/btm369. [DOI] [PubMed] [Google Scholar]
- 27.Kim SY, Volsky DJ. PAGE: parametric analysis of gene set enrichment. BMC Bioinformatics. 2005;6:144. doi: 10.1186/1471-2105-6-144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hulsegge I, Kommadath A, Smits MA. Globaltest and GOEAST: two different approaches for Gene Ontology analysis. BMC Proc. 2009;3(Suppl 4):S10. doi: 10.1186/1753-6561-3-S4-S10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Song S, Black MA. Microarray-based gene set analysis: a comparison of current methods. BMC Bioinformatics. 2008;9:502. doi: 10.1186/1471-2105-9-502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu Q, Dinu I, Adewale AJ, Potter JD, Yasui Y. Comparative evaluation of gene-set analysis methods. BMC Bioinformatics. 2007;8:431. doi: 10.1186/1471-2105-8-431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. Royal Stat. Soc. B. 1995;57:289–300. [Google Scholar]
- 32.Xia J, Psychogios N, Young N, Wishart DS. MetaboAnalyst: a web server for metabolomic data analysis and interpretation. Nucleic Acids Res. 2009;37:W652–W660. doi: 10.1093/nar/gkp356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Neuweger H, Albaum SP, Dondrup M, Persicke M, Watt T, Niehaus K, Stoye J, Goesmann A. MeltDB: a software platform for the analysis and integration of metabolomics experiment data. Bioinformatics. 2008;24:2726–2732. doi: 10.1093/bioinformatics/btn452. [DOI] [PubMed] [Google Scholar]
- 34.Sansone SA, Fan T, Goodacre R, Griffin JL, Hardy NW, Kaddurah-Daouk R, Kristal BS, Lindon J, Mendes P, Morrison N, et al. The metabolomics standards initiative. Nat. Biotechnol. 2007;25:846–848. doi: 10.1038/nbt0807-846b. [DOI] [PubMed] [Google Scholar]
- 35.Dennis G, Jr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003;4:P3. [PubMed] [Google Scholar]