Abstract
chipD is a web server that facilitates design of DNA oligonucleotide probes for high-density tiling arrays, which can be used in a number of genomic applications such as ChIP-chip or gene-expression profiling. The server implements a probe selection algorithm that takes as an input, in addition to the target sequences, a set of parameters that allow probe design to be tailored to specific applications, protocols or the array manufacturer’s requirements. The algorithm optimizes probes to meet three objectives: (i) probes should be specific; (ii) probes should have similar thermodynamic properties; and (iii) the target sequence coverage should be homogeneous and avoid significant gaps. The output provides in a text format, the list of probe sequences with their genomic locations, targeted strands and hybridization characteristics. chipD has been used successfully to design tiling arrays for bacteria and yeast. chipD is available at http://chipd.uwbacter.org/.
INTRODUCTION
With the rapid growth in available genome sequences, metagenomic data sets and the low cost of DNA synthesis, oligonucleotide arrays are now a ubiquitous tool for molecular biologists and others. High-density arrays have been used to monitor mRNA expression levels, determine transcript boundaries, locate protein–DNA interactions, determine DNA methylation patterns or perform comparative genomic hybridization experiments (1,2). Because of the recent advances in array synthesis, arrays can be synthesized on-demand, in small batches, and with no upfront cost, allowing for a greater diversity in targeted organisms and array designs. In addition, the increasing density of unique probes on arrays results in a better coverage of targeted regions of genomes, or complete coverage of small genomes, such as those of bacteria and yeast. Nevertheless, the proper design of oligonucleotide probes is critical for good hybridization with target DNA samples, and therefore, will affect the quality of the resulting experimental data (3). For example, probes need to be sequence-specific to reduce cross-hybridization and the thermodynamic properties of all probes needs to be similar across the array to maximize uniform hybridization.
Most existing software programs for designing of oligonucleotide probes focus on a gene expression platform where only a few probes per messenger RNA are necessary and stringent criteria for the selection of probe characteristics can be applied (4–7). Even though it is possible to use some of these programs to design probes for tiling arrays if they allow the specification of spacing between consecutive probes [OligoWiz 2.0 for example (7)], these programs require additional file manipulations that may be an obstacle for researchers that are less computationally proficient. A few algorithms have been developed for designing tiling arrays, but they focused mainly on solving the problem caused by repeated sequences in very large genomes (8–10); only OligoTiler is accessible as a web tool (http://tiling.gersteinlab.org/OligoTiler/oligotiler.cgi) (11). However, the OligoTiler algorithm does not take into account sequence composition or oligonucleotide length to design probes with similar thermodynamic characteristics. We refer the reader to previous extensive reviews of existing software programs for the design of oligonucleotide probes for additional details (12,13). Here, we present chipD, a public domain and flexible web tool that facilitates the rapid design of specific and thermodynamically similar probes for high-density tiling microarrays. Two different models used to calculate the melting temperature of oligonucleotides have been implemented in chipD to improve the accuracy of melting temperature predictions when designing short (∼25 bp) or long (∼50 bp) oligonucleotide probes. The algorithm used by chipD takes the user DNA sequence and a set of user-specified constraints. The server returns a list of optimized probes that can be submitted to array manufacturers. chipD has been designed to avoid large gaps in the coverage of the target sequence that often result from stringent probe selection criteria. chipD also includes options to design probes for expression microarrays. (chipD is accessible at http://chipd.uwbacter.org/).
INTERFACE
Two options are available in chipD for designing oligos: ‘Genome chip’ and ‘Expression chip’. The ‘Genome chip’ option sets the algorithm to design oligonucleotide probes that tile the entire genomic sequence provided by the user on both strands of the DNA. The ‘Expression chip’ option is used to design probes representing only regions of genomes known or predicted to be transcribed. For ‘Expression chip’, the user must supply only the sequences of the coding strand of the transcribed regions of the genome. The underlying method for probe selection is identical for both options.
Input
Users upload a single sequence file in FASTA format (single or multiple sequences) to the server. Users can use default parameters or specify parameters to tailor probe characteristics to their specific protocol or manufacturer’s specifications. Users can also specify the maximum number of probes returned by the algorithm to accommodate different array formats. Based on the sequence length and the desired number of probes, the algorithm calculates the optimal spacing between probes to obtain uniform coverage of the sequence. For ‘Expression chip’, the algorithm guarantees that each predicted coding region is represented by at least one probe regardless of the coding region predicted length. Next, users can specify an ideal probe length and the desired target-melting temperature for probes. These parameters should be chosen according to the array manufacturer specifications and the users’ individual hybridization protocol. Improperly set melting temperature will result in poor hybridization or cross-hybridization with target DNA, respectively. chipD offers the choice of two alternative models for calculating the melting temperature of oligonucleotides: the nearest neighbor thermodynamic model proposed by SantaLucia (14), which is most accurate for short oligonucleotides (10–30 bp), and an approximate model proposed by Wetmur (15), which performs better for longer oligonucleotides (>40 bp). After selecting a melting temperature model, users can choose to let the algorithm estimate an optimal target-melting temperature using probes whose lengths are determined by the ‘Ideal length’ parameter, and that are randomly selected throughout the input sequence. In our experience with two-color hybridization experiments, longer probes are desirable even if their calculated melting temperatures are higher than specified by the protocol because the resulting signal is more consistent. This is particularly relevant for DNA sequences with a high GC content (see the ‘Example’ section for details). Users can also specify a lower and upper bound for probe lengths, if desired. However, the technical upper limit for probe length is set by the maximum number of cycles the manufacturers use when synthesizing oligonucleotides. Therefore, users are encouraged to specify a maximum number of cycles in chipD instead of limits on probe length. Moreover, restricting the range of possible probe lengths may prevent the algorithm from adequately optimizing probe thermodynamic characteristics and specificity.
Once the parameters are set, the user submits the job, which is queued to the server. Users are given a link where the file containing the selected probes and summary statistics about probe characteristics may be downloaded when the job is completed. Users can use the server anonymously or provide an email address if they wish to receive email notification upon completion of the job. It takes ∼20 min to complete the design of a tiling array for a typical bacterial genome of ∼4 Mbp. Finally, users can easily re-submit jobs if design parameters need to be tweaked.
Output
The output returned to the user is a tabulated text file containing probe sequences along with unique identification strings, the input sequences, the strand orientation and coordinates of the probe sequences, the predicted melting temperature of the probes and their calculated quality scores. Multiple jobs can be submitted to the queue by users. Sequence and results files are stored on the server for a few months and can be accessed later.
ALGORITHM
The scoring of probes is done in two steps. First, all scaffold sequences from the FASTA file are scanned and a global count (based on all scaffolds) is made of each unique sequence of 15 bases and its reverse complement. The global count is then used to define a ‘frequency’ score for each 15 mer. Independently, a ‘complexity’ score is assigned to each 15-mer sequence based on the information content of the sequence. For example, the sequence AAAAAGGGGGCCCCC is less complex than AAGTGATTAGCGTCA. The ‘frequency’ and ‘complexity’ measures are added together to obtain a ‘uniqueness’ score for each 15 mer. The properties of the 15 mer will later be used to determine the overall score of the longer, final probes.
Second, at each position of the input sequence a candidate probe is determined according to a scoring scheme integrating the user-selected input constraints and the sequence quality. At each position, the algorithm iterates over the range of allowed probe lengths (40–70 bases by default) and extracts the probe sequence. For each length, the number of cycles necessary to synthesize the probe is calculated and the iteration stops if this number exceeds the limit (148 cycles by default). The score of each probe is then calculated by summing three components with appropriate weights: (i) a specificity measure; (ii) a target-melting temperature function; and (iii) a target length function. The specificity measure is calculated by summing the ‘uniqueness’ scores of all the overlapping 15 mers that compose the probe according to a weighting function that gives highest weight to 15 mer at the center of the probe and symmetrically less weight toward both ends. The target-melting temperature function calculates how far the predicted melting temperature of the probe deviates from the ideal target temperature determined by the algorithm or set by users. A penalty is applied if the calculated probe melting temperature falls below the target to avoid probes with poor hybridization stability. The target length function slightly penalizes probes with lengths deviating from the ideal probe length as a way to smooth the overall scoring function. After testing all allowed lengths at a position, the algorithm picks and saves the candidate probe with the best score to represent that sequence position. Once the algorithm has scanned all the sequences, every position in the sequences is represented by one candidate probe. The details of the actual scoring function can be found in the reference manual available online.
The final list of probes is selected by the algorithm in an iterative manner. First, all candidate probes are ordered by scores. The probe with the best score is picked and all neighboring probes on the target sequence (based on the spacing constraint) are removed from the list. The process is repeated until the list of candidate probes is exhausted. This algorithm ensures that target sequences are uniformly represented by probes with no significant gaps in the coverage. Finally, if the user is designing a tiling array, the reverse complement of every other probe in the sequence is computed so both strands of DNA are represented on the array. General statistics about the final list of probes are calculated and output for the user.
EXAMPLE: TILING ARRAY DESIGN FOR RHODOBACTER SPHAEROIDES
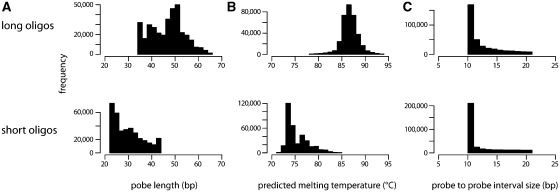
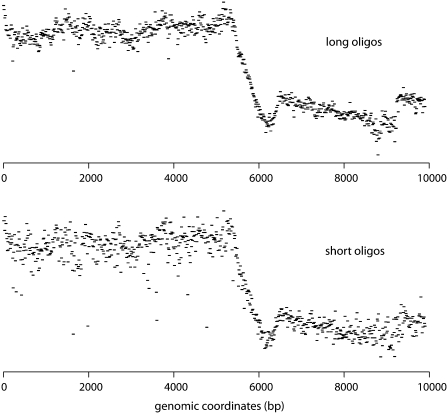
chipD has been successfully used to design tiling arrays for several bacterial and yeast strains (16,17) (N.T. Perna and J.D. Glasner, unpublished data). To illustrate the quality of the probe design, we present data obtained from an experiment with the photosynthetic bacterium R. sphaeroides. Specifically, the interaction of RNA polymerase with DNA was tested by immuno-precipitation and hybridization on custom genome tiling arrays (ChIP-chip experiment). Because R. sphaeroides has a high guanine + cytosine content (70%) in its genome, we wanted to test the effect of probe length or probe melting temperature on the quality of hybridization. To do so, we designed two different sets of probes for custom arrays. The first set, designated ‘short oligos’, was computed using chipD by using the nearest neighbor melting temperature model, setting the target-melting temperature to 74°C and restricting probe lengths to between 22 and 45 bases. The second set, designated ‘long oligos’, was computed by using the approximate melting temperature model, setting the target-melting temperature to 86°C and restricting probe lengths to between 35 and 65 bases. The length, predicted melting temperature and consecutive probe spacing distributions of both sets of probes are represented in Figure 1. There were ∼380 000 unique probes per set, resulting in a full coverage of the R. sphaeroides genome (∼4.6 Mbp) by overlapping probes spaced, on average, every 11 bp relative to the start of each probe. Both designs covered the sequence completely without gaps. Two customs arrays were then synthesized with the two sets of probes. Identical DNA samples labeled with two-colors (immuno-precipitated DNA and genomic DNA control) were hybridized on both arrays in order to compare the difference in design. Figure 2 represents the two sets of signals obtained across a portion of the ribosomal RNA operon in the R. sphaeroides genome. Even though the overall signal is comparable between the two designs, the ‘short oligo’ design produced a significantly higher level of noise. Therefore, from this analysis, we concluded that longer probes have better hybridization quality even though the average melting temperature of the probe set is higher than recommended. The ‘long oligos’ probe set was successfully used to determine the distribution of RNA polymerase and two different σ factor subunits of RNA polymerase on the R. sphaeroides genome in previously published work (16). In addition, another probe set was designed with the same goals to tile the Escherichia coli genome and successfully used to determine the effect of the Rho protein on the distribution of RNA polymerase on the genome (17).
Figure 1.
Histograms representing (A) the distributions of probe lengths; (B) predicted melting temperature; (C) and spacing between consecutive probes for two different oligonucleotide tiling arrays, ‘long oligos’ and ‘short oligos’, used for a ChIP-chip experiment with antibody against R. sphaeroides RNA polymerase. The range of permitted probe lengths was 22–45 bp for the ‘short oligos’ design and 35–65 bp for the ‘long oligos’ design.
Figure 2.
Plots illustrating the difference in signal quality obtained from the hybridization of identical DNA samples on custom arrays containing either the ‘long oligos’ set (top) or the ‘short oligos’ set (bottom). The data represents the relative enrichment signal across a portion of the ribosomal RNA operon resulting from the chromatin immuno-precipitation of RNA polymerase in R. sphaeroides.
CONCLUSIONS
chipD is a public-domain, web-based tool designed to simplify the selection of specific and thermodynamically uniform oligonucleotide probes for high-density tiling array platforms. The server can accommodate relatively small genomes or portions of larger genomes. chipD allows users to specify key parameters, such as desired probe lengths, melting temperature and probe spacing so that probe design can be tailored to users’ specific applications, protocols or platforms. On the other hand, users can also let chipD calculate optimal target-melting temperature or probe spacing to obtain similar thermodynamic characteristics and provide a uniform coverage of the target sequence. A philosophy adopted in chipD that diverges from other probe selection tools is the absence of pre-determined fixed thresholds, which often prevent the selection of probes in difficult sequence regions. Instead, chipD computes the best possible probes across the entire sequence, ensuring full coverage of even the most difficult regions. Users are also provided with probe quality scores and can decide to perform a quality control post-experiment to discard the signals associated with low-quality probes. Finally, in our experience, longer probes appear to give a signal of better quality over shorter probes and should be an important criterion to consider.
FUNDING
Department of Energy Genome to Life BACTER (grants ER63232-1018220-0007203 and DE-FG02-05ER15653 to Y.S.D.); UW-Madison College of Agricultural and Life Sciences (Wisconsin Distinguished Graduate Fellowship to Y.S.D.); UW-Madison Department of Bacteriology (William H. Peterson Predoctoral Fellowship to Y.S.D.); National Institutes of Health (R01-GM62994 to N.T.P., A.J.T.); National Institutes of Health (GM075273 to T.J.D.); UW-Madison Draper Technology Innovation Fund (to T.J.D.). Funding for open access charge: National Institutes of Health (GM075273).
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We thank Madeline Fisher for her help in editing the article.
REFERENCES
- 1.Liu XS. Getting started in tiling microarray analysis. PLoS Comput. Biol. 2007;3:1842–1844. doi: 10.1371/journal.pcbi.0030183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mockler TC, Chan S, Sundaresan A, Chen H, Jacobsen SE, Ecker JR. Applications of DNA tiling arrays for whole-genome analysis. Genomics. 2005;85:1–15. doi: 10.1016/j.ygeno.2004.10.005. [DOI] [PubMed] [Google Scholar]
- 3.Mei R, Hubbell E, Bekiranov S, Mittmann M, Christians FC, Shen MM, Lu G, Fang J, Liu WM, Ryder T, et al. Probe selection for high-density oligonucleotide arrays. Proc. Natl Acad. Sci. USA. 2003;100:11237–11242. doi: 10.1073/pnas.1534744100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chou HH, Hsia AP, Mooney DL, Schnable PS. Picky: oligo microarray design for large genomes. Bioinformatics. 2004;20:2893–2902. doi: 10.1093/bioinformatics/bth347. [DOI] [PubMed] [Google Scholar]
- 5.Li X, He Z, Zhou J. Selection of optimal oligonucleotide probes for microarrays using multiple criteria, global alignment and parameter estimation. Nucleic Acids Res. 2005;33:6114–6123. doi: 10.1093/nar/gki914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rouillard JM, Zuker M, Gulari E. OligoArray 2.0: design of oligonucleotide probes for DNA microarrays using a thermodynamic approach. Nucleic Acids Res. 2003;31:3057–3062. doi: 10.1093/nar/gkg426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wernersson R, Nielsen HB. OligoWiz 2.0–integrating sequence feature annotation into the design of microarray probes. Nucleic Acids Res. 2005;33:W611–W615. doi: 10.1093/nar/gki399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ryder E, Jackson R, Ferguson-Smith A, Russell S. MAMMOT–a set of tools for the design, management and visualization of genomic tiling arrays. Bioinformatics. 2006;22:883–884. doi: 10.1093/bioinformatics/btl031. [DOI] [PubMed] [Google Scholar]
- 9.Lipson D, Yakhini Z, Aumann Y. Optimization of probe coverage for high-resolution oligonucleotide aCGH. Bioinformatics. 2007;23:e77–e83. doi: 10.1093/bioinformatics/btl316. [DOI] [PubMed] [Google Scholar]
- 10.Graf S, Nielsen FG, Kurtz S, Huynen MA, Birney E, Stunnenberg H, Flicek P. Optimized design and assessment of whole genome tiling arrays. Bioinformatics. 2007;23:i195–i204. doi: 10.1093/bioinformatics/btm200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bertone P, Trifonov V, Rozowsky JS, Schubert F, Emanuelsson O, Karro J, Kao MY, Snyder M, Gerstein M. Design optimization methods for genomic DNA tiling arrays. Genome Res. 2006;16:271–281. doi: 10.1101/gr.4452906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lemoine S, Combes F, Le Crom S. An evaluation of custom microarray applications: the oligonucleotide design challenge. Nucleic Acids Res. 2009;37:1726–1739. doi: 10.1093/nar/gkp053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wernersson R, Juncker AS, Nielsen HB. Probe selection for DNA microarrays using OligoWiz. Nat. Protoc. 2007;2:2677–2691. doi: 10.1038/nprot.2007.370. [DOI] [PubMed] [Google Scholar]
- 14.SantaLucia J., Jr A unified view of polymer, dumbbell, and oligonucleotide DNA nearest-neighbor thermodynamics. Proc. Natl Acad. Sci. USA. 1998;95:1460–1465. doi: 10.1073/pnas.95.4.1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wetmur JG. DNA probes: applications of the principles of nucleic acid hybridization. Crit. Rev. Biochem. Mol. Biol. 1991;26:227–259. doi: 10.3109/10409239109114069. [DOI] [PubMed] [Google Scholar]
- 16.Dufour YS, Landick R, Donohue TJ. Organization and evolution of the biological response to singlet oxygen stress. J. Mol. Biol. 2008;383:713–730. doi: 10.1016/j.jmb.2008.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Peters JM, Mooney RA, Kuan PF, Rowland JL, Keles S, Landick R. Rho directs widespread termination of intragenic and stable RNA transcription. Proc. Natl Acad. Sci. USA. 2009;106:15406–15411. doi: 10.1073/pnas.0903846106. [DOI] [PMC free article] [PubMed] [Google Scholar]