Abstract
Site-specific recombinases have become a resourceful tool for genome engineering, allowing sophisticated in vivo DNA modifications and rearrangements, including the precise removal of integrated retroviruses from host genomes. In a recent study, a mutant form of Cre recombinase has been used to excise the provirus of a specific HIV-1 strain from the human genome. To achieve provirus excision, the Cre recombinase had to be evolved to recombine an asymmetric locus of recombination (lox)-like sequence present in the long terminal repeat (LTR) regions of a HIV-1 strain. One pre-requisite for this type of work is the identification of degenerate lox-like sites in genomic sequences. Given their nature—two inverted repeats flanking a spacer of variable length—existing search tools like BLAST or RepeatMasker perform poorly. To address this lack of available algorithms, we have developed the web-server SeLOX, which can identify degenerate lox-like sites within genomic sequences. SeLOX calculates a position weight matrix based on lox-like sequences, which is used to search genomic sequences. For computational efficiency, we transform sequences into binary space, which allows us to use a bit-wise AND Boolean operator for comparisons. Next to finding lox-like sites for Cre type recombinases in HIV LTR sequences, we have used SeLOX to identify lox-like sites in HIV LTRs for six yeast recombinases. We finally demonstrate the general usefulness of SeLOX in identifying lox-like sequences in large genomes by searching Cre type recombination sites in the entire human genome. SeLOX is freely available at http://selox.mpi-cbg.de/cgi-bin/selox/index.
INTRODUCTION
Site-specific recombination constitutes an important part of the functional genomics toolbox allowing for highly specific and efficient targeted genomics studies (1). The instruments of this engineering technology are site-specific recombinases (SSRs) that bind to particular DNA target sequences consisting of two inverted repeats flanking a spacer of variable length (2). The best-known SSR is the Cre/lox system from bacteriophage P1, where the Cre recombinase catalyzes the exchange of DNA fragments that are flanked by so-called loxP sites (3). SSRs of the Cre/lox type have meanwhile become a part of the standard repertoire for genome engineering in knock-out and knock-in studies in both, plant and animal systems.
One limitation of the SSR systems is that the recognition sequence has to be introduced into the genome of the organism before the recombinase can be usefully employed. To overcome this limitation considerable efforts have focused on adapting recombinases to recognize pre-defined sequences (4). One successful application of this kind has been the development of the Tre recombinase (5). To derive Tre, Cre-recombinase was evolved in vitro using a technique called substrate-linked protein evolution [SLiPE, (4)] to recognize the lox-like long terminal repeat (LTR) sequences of a specific HIV-1 subtype. In this proof-of-principle study, the LTR regions with closest similarity to a loxP site of an HIV-1 subtype were used to select for enzymes that were mutated using several cycles of directed evolution. The resulting enzyme, Tre, was able to excise the HI provirus from the host genome by recognizing an asymmetric sequence present in the 3′ and 5′LTRs. The work of Sarkar et al. (5) demonstrates that it is feasible to engineer SSRs that have altered binding specificities and can tolerate mismatches within, as well as between lox-like flanking regions.
In order to identify a suitable HIV strain for directed evolution of Cre, manual comparisons of HIV LTR sequences to the Cre loxP site were carried out. Alignments of LTR sequences with the lox site were manually constructed and compared to each other, which turned out to be a time-consuming and also error-prone procedure. In order to find the most suitable lox-like site, one would ideally use a mixture of lox-like sites such as loxP, loxH or rox to search degenerate motifs, which are recognized by highly similar, yet not identical recombination systems. Given the high similarity of these recombinases (6), it is likely that hybrid recombination systems could be generated by family shuffling (7), resulting in enzymes with altered target specificity. A second argument towards a completely automated search strategy is that a manual approach will not provide any information of achievable integration specificity, as it precludes searches against entire genomes for endogenous lox-like sites that could be unintentionally targeted. Likewise, no efforts have so far been made to computationally assess the presence of lox-like target sites of existing, non-standard recombination systems in entire genomes, which could be adapted for genome engineering in model organisms.
In all cases, what is needed is an automated way to identify degenerate lox-like sites in genomic sequences that are not well characterized. Known recognition sites of recombinases, which have been described in literature, can be used as a query for such a search against a genomic stretch. Given the nature of such regions, the use of standard sequence similarity search tools is precluded. Using a manual approach, one recombination site was singled out in the study of Sarkar et al. (5), yielding a recombinase Tre with high-strain specificity. If recombinases, however, are to be used as therapeutic agents against retroviral genomes, it is critical to engineer recombinases to target recognition sites present across as many strains of the retrovirus as possible. As BLAST does not perform well with such short redundant sequences, we considered using HMMER (8), RepeatMasker (9) or the palindrome program from the Emboss suite of packages (10). HMMER was developed to find remote homologs based on a probabilistic model of a sequence pattern being looked for, which is not the nature of the search we intended to perform. HMMER could be forced to address our search question, but the amount of pre- and post-processing of data and parameters would make it highly error-prone and inefficient. RepeatMasker only looks for repeats and low-complexity regions that are already well characterized, which is again not the nature of our search. The Emboss palindrome program comes closest to addressing our search question, but does not allow for a search to be defined, but rather results in a list of possibilities of lox-like sites. These we would then have to match to the lox-site signature we are interested in. This would make the search convoluted and cumbersome. Finally, another possibility would have been to work with Stochastic Context-Free Grammars (SCFGs), which have been used extensively for detecting RNA molecules in genomes and which have become especially popular in the field of RNA secondary structure prediction (11,12). In order to apply SCFGs to our problem, we would, however, have to know the nature, as well as the frequency of individual mutations of different LTRs, both of which are currently not well defined. In the case of the Cre SSR, we can, for instance, only use the loxP, loxH and with restraints the rox sites for constructing an evolutionary model. The lack of available sequences would lead to a poorly described probability model for subsequent identification of more distantly related lox-like sites in genomic sequences.
To address the lack of a program and a method to find degenerate lox-like sites, we have developed the web server SeLOX, which provides an easy to use interface for searching degenerate lox-like sites in genomic sequences. To make the search computationally efficient, we decided to use binary operations on the sequences after they are transformed into bit strings.
WEB SERVER WORKFLOW
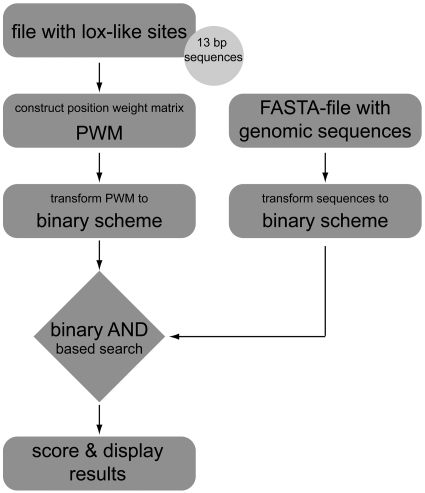
The workflow (Figure 1) can be divided in three steps and is described below.
-
First, a position weight matrix for the flanking regions is constructed based on known recognition sites of a recombinase. The first page of the web server asks for the user to upload a text file containing the left-flanking regions of lox-like sites that the recombinase under study recognizes. The file should contain sequence regions of identical length, which is used to initialize the horizontal dimension of the position weight matrix for both, the left- and right-flanking regions. The content of an example file of lox-like flanking region is shown below:
ATAACTTCGTATA
ATATATACGTATA
TAACTTTAAATAA
The web server calculates and displays a position weight matrix corresponding to the flanking region sequences that were submitted (Figure 2A). In brief, based on the frequency of each of the 4 nt being present at a particular position in the lox-like flanking regions submitted, the weight matrix captures the probability of a certain base being present at that position (‘Methods’ section).
Next, a file containing the FASTA-formatted genomic sequences that is to be searched for the specified lox-like sites has to be provided. In this step, the user also sets the various parameters for the search: the length of the spacer and the tolerance thereof, the number of acceptable mismatches to the position weight matrix and the asymmetry that can be tolerated between the left- and right-flanking regions.
Finally, the web server performs a binary search for lox-like sites in the user-provided sequences and scores the resulting hits based on the initial weight matrix (‘Methods’ section). The web server displays the position weight matrix that was used for the search as a sequence logo implemented using HTML and in-line CSS. For each lox-like site found, the web server shows its sequence, the number of sequences containing this particular lox-like site and the score computed for the lox-like site (Figure 2B). The output is grouped and sorted according to the number of sequences containing a particular lox-like site, and the score. For convenience, the web server also marks those nucleotides in the lox-like site which are mismatches relative to the queried position weight matrix (Figure 2B, red dots) and those nucleotides in the right-flanking region, which are asymmetric i.e. not reverse-complementary to the corresponding nucleotides in the left flanking region (Figure 2B, orange dots).
Figure 1.
Workflow of the SeLOX web server. The user-provided lox-like sites and genomic sequences are transformed into the binary space and compared using the binary AND operation. The resulting lox-like hits are scored and displayed to the user.
Figure 2.
(A) First result page of the SeLOX web server showing the position weight matrix of the submitted lox-like sites. The user next needs to upload the genomic sequence stretches to be searched. (B) Results of the SeLOX server using the sample data files. The position weight matrix for the submitted lox-like sites is shown on top of the window, followed by the number and detailed display of lox-like sites found. Mismatches within a flanking regions are marked by red dots above the sequence, those that break symmetry by orange dots below.
METHODS
The web server is implemented entirely in Python v2.4, and uses the cgi standard python library module for interaction between the web pages and the analysis.
Bit-representation of nucleotides
While the rationale of most sequence similarity analysis tools like BLAST and other motif recognitions tools is the homology between sequences, the search for lox-like sites does not have this basis, and is a pure string search question. To allow for the repetitive nature of the lox-like sites, and to make the search as efficient as possible, we decided to exploit bit-wise operations after transforming the sequences into integers.
The principal idea underlying SeLOX is the transformation of nucleotides into binary space, which are then stored as integers corresponding to the binary transformations. A 4-bit string is needed to represent all possible combinations of nucleotides at a given position of a sequence region. We follow the scheme A=1000, T=0100, G=0010, C=0001 to represent each of the nucleotides. For instance, to account for nucleotides A, G and C at a particular position, the corresponding transformation in binary space would be 1011, which would be stored as the integer 13.
Transformation of the input sites into a query bit string vector
SeLOX takes as input a set of flanking regions of lox-like sites. It then builds a position weighted matrix of this set, where corresponding to each position, every nucleotide has a weight equal to its frequency at that position in the provided lox-like sites. Each position in the query is then transformed into a 4-bit string using the scheme described earlier, thus creating one part of the lox-like query vector. To search for the reverse complement of the input regions, we perform the same procedure by converting the nucleotides represented in the position-weighted matrix into their reverse-complements and inverting the position weighted matrix. This results in the lox-like query vector that is used for the search.
Search procedure
Using the scheme described earlier, SeLOX transforms all individual sequences in the uploaded FASTA file into an integer vector, where each element of the vector is representative of the binary transformation of the nucleotide at that position in the sequence. Cassettes of elements that are of the length of the lox-like sites from the sequence vector are then used for the search using a binary AND operation between these cassettes and the query vector. A non-zero integer results if there is a match between the query and the sequence vector at a particular position, a zero integer when there is no match.
For example, if at a particular position, we have nucleotides [A, G] in the query sequence, and an A in the sequence being searched, the transformations of the query and the sequence searched would be 1010 and 1000, respectively. The binary AND operation on 1010 and 1000 yields 1000 indicating that there is a match, and also a readout as to which nucleotide matched. For each cassette of the sequence vector, the number of mismatches is simply the count of 0 in the vector resulting from the binary AND. A match is found if the number of mismatches is less than or equal to the number of mismatches specified by the user in the web interface.
A result vector with the length of the uploaded lox-like sites, holds the results of each cassette that correspond to the starting position of the cassette in the analyzed sequence. Using different identifiers for the forward and the reverse query vectors from the lox-like input sequences, each cassette is marked with 1 if there is a match to the forward vector, a 2 if there is a match to the reverse vector and 3 if there is a match to both the forward and reverse vectors. A 0 marks no match to either the forward or reverse query vectors.
We perform a linear walk starting from the beginning of the result vector, looking for a 1 or a 3 indicating the presence of a match to the forward query vector. Next, we look for a subsequent 2 or 3 identifier, present within a region of the result vector defined by the spacer length specified by the user. We check for asymmetry between the forward and the reverse-flanking regions, and check if the number of positions which are asymmetric is less than or equal to the maximum asymmetry limit set by the user. When such a region is found, we have successfully identified a lox-like site corresponding to the input lox-like regions in the input sequences.
Scoring
We perform a naïve scoring of each lox-like hit, using the weights calculated as described earlier. A match at a position results in a positive score equal to the weight for the nucleotide matched at that position from the position weighted matrix, and a mismatch at a position yields a negative score of the maximum weight derived for that particular position. The sum of the scores at all positions gives a score indicative of the match to the query lox-like site being looked for. Primarily, only the mutations in the left flanking sequence are scored. Mutations in the right repeat are only indirectly scored by the positions that break the symmetry. The score is primarily used to sort hits, whereby the primary decisive factor for sorting is the number of strains a lox-like site is found in.
APPLICATIONS
Identification of Cre family lox-like sequences in HIV-LTRs
SSRs have recently been used to remove the HIV-1 provirus by recombination via targeted sites, loxLTR, present in the LTR regions flanking the provirus (5). The manual comparison of loxP to LTR sites singled out the LTR sequence of a specific HIV-1 strain (TZB0003). Ideally, an engineered recombinase that was designed for therapeutic purposes against retroviruses like HIV would target a broader spectrum of strains. In order to achieve this form of strain-agnostic agent, a rather degenerate site would have to be chosen for directed enzyme evolution. A manual approach for the selection of such a strain-agnostic sequence would not be doable. In order to test the performance of SeLOX to identify potentially strain-agnostic lox-like sequences, we used loxP, loxH and rox sites (Supplementary File S1) as input for SeLOX and searched against the sequence space of eight different HIV1-LTR regions (Supplementary File S2). For the search, we allowed up to five mutations at the flanking region and eight mismatches for the symmetry (Supplementary Figure S1). Among the identified sequences, we found the initial loxLTR site used in Sarkar et al. (5). SeLOX identified this sequence as a high-scoring (score=6) lox-like site not shared by the other strains. In addition to this original site, SeLOX also identified an agnostic loxLTR sequence that was present in six of the eight strains with four mutations in the flanking region and eight mismatches in symmetry (score=5.32). This sequence would be a possible next choice for usage in directed SLiPE of Cre family recombinases. Our results further demonstrate the power of SeLOX, which is not only able to identify suitable lox-like sites for directed evolution but also identify them across different viral strains.
Re-use of existing recombinases
Cre is a very popular, however not the only SSR currently in use. Other recombination systems that have been extensively used are for example Flp (13) or phiC31 (14). A number of Flp-related recombinases exist from other yeast strains that recognize different, but related target sequences (15). Flp-family recombinases present an attractive alternative to the Cre-family recombinases for the development of enzymes with desired target specificities. As proof of principle, we used recognition target sites of six yeast recombinases (Supplementary File S1) as input for SeLOX and searched for similar sites in the eight HIV-1 LTR regions described earlier. As the nature of the six yeast recombinases is quite divergent, SeLOX found >2700 lox-like sites in the eight HIV LTRs when we allowed five mutations in the flanking region and eight mismatches in symmetry (data not shown). When we limited the lox sites to the three more conserved target sites of the recombinases KW, SM1 and SB2 using the same settings, we identified 28 loxLTR sites, whereby the most agnostic variant is found in six of the eight HIV strains (Supplementary Figure S2, top hit). In summary, our data suggests that some of the yeast recombinases might be equally suitable for directed enzyme evolution and therefore, as therapeutic agents to remove retroviruses from infected hosts.
Identification of lox-like sites in vertebrate genomes
As SeLOX is computationally very efficient, it can also be used to identify any divergent, lox-like site in entire genomes. This would be especially useful to ensure specificity when designing SSRs using SLiPE. As a proof of principle, we have applied SeLOX to find degenerate lox-like sequences of the loxP type in the human genome. So far, this question has only been addressed experimentally (16). We used the loxP/loxH and rox sequences described earlier and allowed two mismatches in the flanking region, as well as for symmetry. After removing consecutive di-nucleotide repeats from the hitlist, we found a total of 229 lox-like sites in the human genome (Supplementary File S3). Interestingly, the most conserved site that was identified by SeLOX is located on Chromosome 10 with a very high score of 15.36, 36 and a spacer of 7 nt (sequence ATACCTTCGTGTA GGAGCCC TACACGAAGGTAT). This lox-like site is a perfect, inverted repeat and has a single mismatch compared to the original loxP site. Given the high similarity of this sequence to the original loxP site, it could very well be that this site is a direct target of Cre itself, if the enzyme could tolerate a 7-nt spacer. Alternatively, it should be relatively straightforward to develop a recombinase targeting this sequence by SLiPE, which may then be useful to deliver recombinase-mediated DNA into this locus via site-specific integration.
CONCLUSIONS AND PERSPECTIVES
We have developed SeLOX, an easy to use web server designed for identification of lox-like sites for genomic recombination studies. SeLOX finds its use both in functional genomics studies and in therapeutic applications like removal of host genome-integrated retroviruses. One obvious further application would be to identify lox-like sites of endogenous retroviruses in genomes, which could among other applications be interesting medical targets (17–23). So far this aspect has only been addressed experimentally. Our proof of principle search for lox-like sites in the human genome has proven that SeLOX can be used to search lox-like sites within large genomic sequences. In order to identify unknown retroviral recombination sites, average values have to be assigned to the flanking regions and a range of spacer lengths has to be allowed. However, when using such settings, SeLOX is susceptible to detecting highly repetitive, non-lox like elements in genomic DNA and a preceding repeat-filter would have to be implemented to eliminate those sequence features.
SeLOX is, due to its speed, especially interesting for genome-wide searches of degenerate inverted repeats. In this context, it would be helpful to further build on the idea of the binary transformation to make the search computationally even more efficient. Since the binary transformation of the sequences can be treated as a discrete signal wave, we are considering the use of Fourier transforms to perform the matching of the query site pattern against whole genomes. Alternatively, an obvious next step would be to implement the code in C for application in genome-wide searching.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Max Planck Gesellschaft and German Research Foundation (DFG). Funding for open access charge: Max Planck Institute of Molecular Cell Biology and Genetics Pfotenhauerstrasse and DFG (DFG-Grant SFB 655).
Conflict of interest statement. None declared.
Supplementary Material
REFERENCES
- 1.Garcia-Otin AL, Guillou F. Mammalian genome targeting using site-specific recombinases. Front. Biosci. 2006;11:1108–1136. doi: 10.2741/1867. [DOI] [PubMed] [Google Scholar]
- 2.Voziyanov Y, Pathania S, Jayaram M. A general model for site-specific recombination by the integrase family recombinases. Nucleic Acids Res. 1999;27:930–941. doi: 10.1093/nar/27.4.930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Van Duyne GD. A structural view of cre-loxp site-specific recombination. Ann. Rev. Biophys. Biomol. Struct. 2001;30:87–104. doi: 10.1146/annurev.biophys.30.1.87. [DOI] [PubMed] [Google Scholar]
- 4.Buchholz F, Stewart AF. Alteration of Cre recombinase site specificity by substrate-linked protein evolution. Nat. Biotechnol. 2001;19:1047–1052. doi: 10.1038/nbt1101-1047. [DOI] [PubMed] [Google Scholar]
- 5.Sarkar I, Hauber I, Hauber J, Buchholz F. HIV-1 proviral DNA excision using an evolved recombinase. Science. 2007;316:1912–1915. doi: 10.1126/science.1141453. [DOI] [PubMed] [Google Scholar]
- 6.Anastassiadis K, Fu J, Patsch C, Hu S, Weidlich S, Duerschke K, Buchholz F, Edenhofer F, Stewart AF. Dre recombinase, like Cre, is a highly efficient site-specific recombinase in E. coli, mammalian cells and mice. Dis. Model Mech. 2009;2:508–515. doi: 10.1242/dmm.003087. [DOI] [PubMed] [Google Scholar]
- 7.Crameri A, Raillard SA, Bermudez E, Stemmer WP. DNA shuffling of a family of genes from diverse species accelerates directed evolution. Nature. 1998;391:288–291. doi: 10.1038/34663. [DOI] [PubMed] [Google Scholar]
- 8.Eddy SR. Profile hidden Markov models. Bioinformatics. 1998;14:755–763. doi: 10.1093/bioinformatics/14.9.755. [DOI] [PubMed] [Google Scholar]
- 9.Chen N. Using RepeatMasker to identify repetitive elements in genomic sequences. Curr. Protoc. Bioinform. 2004 doi: 10.1002/0471250953.bi0410s05. Chapter 4: Unit 4.10. [DOI] [PubMed] [Google Scholar]
- 10.Rice P, Longden I, Bleasby A. EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet. 2000;16:276–277. doi: 10.1016/s0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]
- 11.Sakakibara Y, Brown M, Hughey R, Mian IS, Sjolander K, Underwood RC, Haussler D. Stochastic context-free grammars for tRNA modeling. Nucleic Acids Res. 1994;22:5112–5120. doi: 10.1093/nar/22.23.5112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bradley RK, Pachter L, Holmes I. Specific alignment of structured RNA: stochastic grammars and sequence annealing. Bioinformatics. 2008;24:2677–2683. doi: 10.1093/bioinformatics/btn495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Golic KG, Lindquist S. The FLP recombinase of yeast catalyzes site-specific recombination in the Drosophila genome. Cell. 1989;59:499–509. doi: 10.1016/0092-8674(89)90033-0. [DOI] [PubMed] [Google Scholar]
- 14.Groth AC, Olivares EC, Thyagarajan B, Calos MP. A phage integrase directs efficient site-specific integration in human cells. Proc. Natl Acad. Sci. USA. 2000;97:5995–6000. doi: 10.1073/pnas.090527097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Blaisonneau J, Sor F, Cheret G, Yarrow D, Fukuhara H. A circular plasmid from the yeast Torulaspora delbrueckii. Plasmid. 1997;38:202–209. doi: 10.1006/plas.1997.1315. [DOI] [PubMed] [Google Scholar]
- 16.Thyagarajan B, Guimaraes MJ, Groth AC, Calos MP. Mammalian genomes contain active recombinase recognition sites. Gene. 2000;244:47–54. doi: 10.1016/s0378-1119(00)00008-1. [DOI] [PubMed] [Google Scholar]
- 17.Huang W, Li S, Hu Y, Yu H, Luo F, Zhang Q, Zhu F. Implication of the env gene of the human endogenous retrovirus W family in the expression of BDNF and DRD3 and development of recent-onset Schizophrenia. Schizophr. Bull. 2010 doi: 10.1093/schbul/sbp166. doi:10.1093/schbul/sbp166 [25 January 2010, Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 19.Jeong BH, Lee YJ, Carp RI, Kim YS. The prevalence of human endogenous retroviruses in cerebrospinal fluids from patients with sporadic Creutzfeldt-Jakob disease. J. Clin. Virol. 47:136–142. doi: 10.1016/j.jcv.2009.11.016. [DOI] [PubMed] [Google Scholar]
- 20.Lower R, Lower J, Kurth R. The viruses in all of us: characteristics and biological significance of human endogenous retrovirus sequences. Proc. Natl Acad. Sci. USA. 1996;93:5177–5184. doi: 10.1073/pnas.93.11.5177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pullmann R, Jr, Bonilla E, Phillips PE, Middleton FA, Perl A. Haplotypes of the HRES-1 endogenous retrovirus are associated with development and disease manifestations of systemic lupus erythematosus. Arthritis Rheum. 2008;58:532–540. doi: 10.1002/art.23161. [DOI] [PubMed] [Google Scholar]
- 22.Perron H, Lang A. The human endogenous retrovirus link between genes and environment in multiple sclerosis and in multifactorial diseases associating neuroinflammation. Clin. Rev. Allergy Immunol. 2009 doi: 10.1007/s12016-009-8170-x. [21 August 2009, Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 23.Serafino A, Balestrieri E, Pierimarchi P, Matteucci C, Moroni G, Oricchio E, Rasi G, Mastino A, Spadafora C, Garaci E, et al. The activation of human endogenous retrovirus K (HERV-K) is implicated in melanoma cell malignant transformation. Exp. Cell Res. 2009;315:849–862. doi: 10.1016/j.yexcr.2008.12.023. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.