Abstract
Molecular simulations offer important mechanistic and functional clues in studies of proteins and other macromolecules. However, interpreting the results of such simulations increasingly requires tools that can combine information from multiple structural databases and other web resources, and provide highly integrated and versatile analysis tools. Here, we present a new web server that integrates high-quality animation of molecular motion (MM) with structural and functional analysis of macromolecules. The new tool, dubbed POLYVIEW-MM, enables animation of trajectories generated by molecular dynamics and related simulation techniques, as well as visualization of alternative conformers, e.g. obtained as a result of protein structure prediction methods or small molecule docking. To facilitate structural analysis, POLYVIEW-MM combines interactive view and analysis of conformational changes using Jmol and its tailored extensions, publication quality animation using PyMol, and customizable 2D summary plots that provide an overview of MM, e.g. in terms of changes in secondary structure states and relative solvent accessibility of individual residues in proteins. Furthermore, POLYVIEW-MM integrates visualization with various structural annotations, including automated mapping of known inter-action sites from structural homologs, mapping of cavities and ligand binding sites, transmembrane regions and protein domains. URL: http://polyview.cchmc.org/conform.html.
INTRODUCTION
A growing number of tools are being developed for visualization of macromolecules and animation of their motion with the goal of facilitating experimental and computational studies of macromolecules and their complexes. Many of these tools are available as stand-alone packages, such as VMD (1), Swiss-PDBViewer (2) or PyMol (3), which combine impressive rendering capability with versatile analyses, scripting languages and extensible architecture. At the same time, web-based tools are being developed to provide easy-to-use intuitive interfaces, often employing platform-independent rendering programs, such as Jmol (http://www.jmol.org/ an open-source Java viewer for chemical structures in 3D). Several resources have specifically been developed to provide animation of MM, including Molecules in Motion (http://www.moleculesinmotion.com/), AISMIG (4), POLYVIEW-3D (5) and Protein Movie Generator (6).
One of the major drawbacks of stand-alone tools is their limited awareness of rich on-line resources and databases. In addition, such tools are often rather difficult to use for researchers with a limited technical expertise in computing. On the other hand, existing on-line tools for MM require further improvements both in terms of the quality of visualization and integration with analysis and annotation tools. Enhanced tools for visualization, annotation and automated analyses that take advantage of ever more complex network of web resources and databases is an emerging need in the fields of structural biology, functional genomics and molecular simulations.
Here, we present a new web-based tool, POLYVIEW-MM, which integrates high-quality animation with structural annotation of MMs, and allows both qualitative and quantitative analysis of the results of macromolecular simulations. To facilitate structural analyses, POLYVIEW-MM combines: (i) interactive view and scripting-based conformational analysis using Jmol and its tailored extensions; (ii) versatile annotation and publication quality animation using PyMol, which is available through POLYVIEW-3D; and (iii) customizable 2D summary plots that provide insights into MM in terms of changes in secondary structure (SS) states, relative solvent accessibility (RSA) of individual residues in proteins, as well as distance plots and analysis of intra- and inter-molecular contacts. The latter type of analysis is facilitated by 2D summary plots highlighting residues involved in complex formation, including two important special cases of interaction interfaces in alternative models/conformers of protein–protein and protein–ligand complexes obtained from protein and small molecule docking simulations, respectively.
At the same time, using easy to generate animations and other graphical representations, POLYVIEW-MM enables visualization of trajectories generated by molecular dynamics (MD) and related simulation techniques. Moreover, alternative conformers of small molecules, proteins and other macromolecules can be visualized using molecular movies. As an input, POLYVIEW-MM accepts files in the standard Protein Data Bank (PDB) format with multiple models. This format has been widely adopted by servers and programs that aim at protein structure prediction and analysis of MM. Examples of the latter include tools for analysis of distortions and slow coordinated motions in proteins, as obtained using, e.g. AD-ENM (7) or MolMovDB (8). To enable direct analysis of MD trajectories, POLYVIEW-MM accepts trajectory files in the DCD format, which is supported by widely used simulation packages NAMD (9) and CHARMM (10), as well as the TRR format supported by GROMACS (11). For the analysis of small molecule docking, the DLG format used by the popular AutoDock program (12) is accepted as well.
POLYVIEW-MM incorporates into the analysis of macromolecular simulations many structural and functional annotations, including automated mapping of protein domains from the Pfam database of protein families (13), membrane domains from the PDB_TM database (14) and protein–protein interaction interfaces from the PDB database (15). The latter option can greatly facilitate structural and functional analyses in the context of constantly growing number of resolved protein complexes. The mapping is conducted using sequence homology search. The results of several annotation tools, including CASTp for the identification of structural cavities (16), and SPPIDER for predicting putative novel interaction sites (17) can be automatically retrieved and mapped into POLYVIEW-MM output as well. Additionally, the server accepts requests for calculation and mapping of evolutionary conservation scores derived from multiple sequence alignment using PSI-BLAST (18). This option enables analysis of conserved functional hotspots in the context of MM.
METHODS
Input and output
As the primary input format, POLYVIEW-MM uses 3D coordinate files in the PDB (15) format that can contain multiple structural models of the same molecule, or a complex of molecules, including proteins, nucleic acids and small molecules. This format can be used with each of the basic types of queries that include: (i) NMR-based ensembles of models (these queries can also be specified by the PDB entry ID); (ii) trajectories obtained using MD simulations; (iii) molecular morphs or snapshots representing slow coordinated motions obtained, e.g. using elastic network models; and (iv) protein–protein or protein–ligand complexes obtained using docking simulations. Other specific input formats include the DCD and TRR formats for MD trajectories and the DLG (AutoDock) format for ligand docking simulations. For other MD formats, utilities similar to CatDCD (1) or MDanalysis (http://code.google.com/p/mdanalysis/) may need to be used to convert MD trajectories to the multiple model PDB or DCD formats. For faster upload of the data, MD trajectories can be submitted using compressed files in ZIP, GZIP and BZIP2 formats.
As part of the output, POLYVIEW-MM allows to display trajectories and multiple conformers using interactive animations and static views in Jmol, coupled with tailored selection and annotation options. These can be subsequently imported into POLYVIEW-3D in order to generate publication quality static pictures and animations using PyMol rendering. POLYVIEW-MM also generates a number of customizable 2D plots that provide simple yet informative summaries of conformational changes and differences between individual models. All movies and plots generated by the server can be deposited with user-defined annotations to an image library as a mechanism to document and share data with colleagues.
Motion at a glance
To provide an overview of conformation changes in proteins, POLYVIEW-MM generates 2D plots that display SS and RSA states for individual residues in each snapshot of a trajectory, or each conformer submitted as a multiple model PDB file. SS and RSA are calculated using the DSSP program (19), using either eight (G, H, I, T, E, B S, and C, as defined in DSSP) or three SS states (H—α-helix, E—β-strand, and C—loop). Surface-exposed area computed using DSSP is normalized by the maximum value of the solvent exposed surface area for a given type of amino acid as determined in (20), resulting in RSA values between 0% and 100%, with the latter corresponding to a fully exposed residue. Real-valued RSA are subsequently projected into 10 discrete states for the purpose of displaying them as shaded boxes, with black boxes corresponding to fully buried and white boxes to fully exposed residues, respectively. At each position, entropy in terms of the SS and RSA states is computed in order to capture conformational variability at that position in a trajectory or a set of conformers. In addition, simple distance plots between interactively selected pairs of atoms or residues can be generated for analysis of molecular trajectories.
Protein–protein and protein–ligand interfaces
Following previous studies (17,21,22), we define protein–protein interaction sites based on the RSA change upon complex formation, i.e. RSA difference between an unbound and bound (complex) structure of an individual chain. The procedure and thresholds used to assign an amino acid residue as an interaction site can be found in (17). Protein–protein interaction interfaces can be characterized in terms of the surface area buried upon complex formation, amino acid properties and the presence of conserved hot spots, facilitating analysis of protein docking simulations, for instance. Protein–ligand contacts are determined using the respective procedure adopted in Protein Explorer (23) and subsequently in the FirstGlance in Jmol server (FGiJ, http://firstglance.jmol.org/) that accounts for hydrogen bonds, water and salt bridges, hydrophobic and aromatic ring interaction and different types of metals binding. For the corresponding bond distance definitions, the reader is referred to the FGiJ documentation. Mapping specific residues in contact with the ligand in alternative poses of protein–ligand complexes can be used to assess the results of docking simulations. Consistency of interacting sites observed in protein–protein or protein–ligand docking models is measured by computing frequency of being in contact with the interacting co-factor.
Structural annotations
POLYVIEW-MM provides the ability to automatically annotate images and movies with structural and functional characteristics derived using annotation and prediction tools. The following structural annotations can be retrieved and mapped at present (more to be included in future): (i) structural cavities determined using the CASTp server (16); (ii) protein domains and motifs annotated in the Pfam database (13); (iii) transmembrane domains identified using the PDB_TM database (14); (iv) known protein–protein interaction interfaces found in complexes deposited to PDB; and (v) putative interaction sites predicted using SPPIDER (17); position-specific evolutionary scores derived from multiple sequence alignments using PSI-BLAST (18). Retrieval and mapping of available structural annotations is conducted using sequence-based homology search using BLAST (24). Homology hits with E-value equal to or lower than 0.001 with sufficient level of sequence identity (70%) are considered when performing searches against PDB, Pfam and PDB_TM databases, and residues within structural and functional hot spots (e.g. interaction interfaces) are mapped into the query structure based on the sequence alignment. Specifically, all homology hits, as defined above, from Pfam and PDB are retrieved for annotation, whereas only the best homology hit is used to map transmembrane domains. Multiple sequence alignment is performed against nr database with E = 0.001 cutoff. Conservation scores are computed from the position-specific scoring matrix obtained after three PSI-BLAST iterations.
EXAMPLES
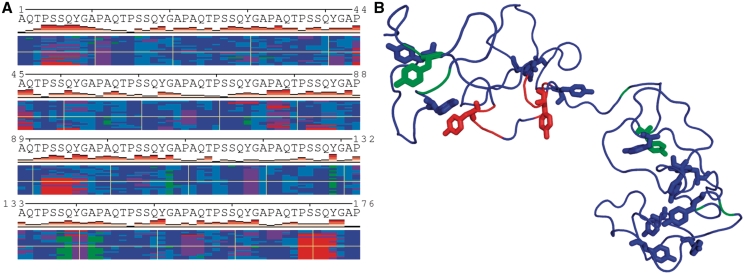
To illustrate the capabilities of POLYVIEW-MM, we present two examples. Since molecular movies cannot be directly displayed here, these examples focus on static pictures and are supplemented by animations in the documentation and image gallery available from the home page. The first example presented here concerns visualization and analysis of MD trajectories. In Figure 1, an NAMD generated trajectory for an idealized resilin-like peptide, AN16 (25), is shown using a 2D plot that characterizes conformational changes in terms of SSs of individual snapshots in the trajectory. The number of times a given residue changes the SS state, which is a measure of conformational transitions at that position throughout the trajectory is represented by white through pink to red bars shown below the amino acid sequence. In addition, one particular 3D snapshot is shown in Figure 1B for illustration of persistent disorder observed in this case (only transient helices and short beta strands are observed in some repeats). This persistent disorder can also be easily discerned using a 2D plot with RSA profiles of each structural snapshot (data not shown). AN16 comprises 16 repeats of an 11-mer derived from elastomeric insect protein called resilin (26), which is characterized by the highest known resilience of all known elastic materials. Upon stretching, entropy of the disordered relaxed state is lost, giving rise to an entropic force (27). In nature, multiple resilin chains are cross-linked at tyrosine residues to form an elastic fiber, which signifies the importance of the distribution of tyrosine pairwise distances that can be generated in POLYVIEW-MM together with movies and other representations of the trajectory.
Figure 1.
Visualization of MD trajectory for resilin-like peptide with changes in SS states along the trajectory shown in (A). Red colors correspond to different helices, green to beta states and blue to unstructured regions, bends and magenta to turns, respectively (see POLYVIEW-MM documentation for details). Each row corresponds to an individual snapshot of the structure, an example of which is shown in (B) with tyrosine side chains shown in colors corresponding to initial SS states and using stick models. Note the lack of regular SSs.
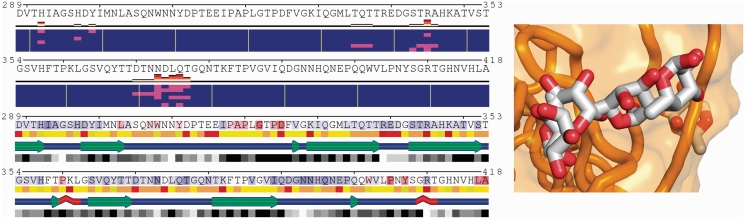
The second example presented here illustrates how POLYVIEW-MM can be used to visualize and analyze docking simulations (Figure 2). Specifically, the widely used package for small molecule docking, AutoDock4 (12), has been used to generate alternative poses (models of the receptor–ligand complexes) for fucose bound to the capsid protein of a specific strain (VA387) of norovirus that causes large outbreaks of gastroenteritis (28). Noroviruses recognize histoblood type antigens that are presented by host cells in the gut (28), and binding of the fucose ring in these polysaccharides to norovirus capsid proteins plays an important role (29). Examples of alternative fucose poses superimposed together are shown in the right panel using a static 3D picture (the corresponding animation is available from the POLYVIEW-MM gallery). The corresponding interaction interfaces are shown in the left upper panel, with residues in contact with the ligand indicated by magenta (each row represents one docking model). In the lower left panel, evolutionary conservation of amino acid residues is indicated by the background of the amino acid letter (red corresponds to highly conserved, and blue to variable positions), chemical profile (with yellow indicating hydrophobic and red and brown hydrophilic positions) is shown below the sequence row, and the SSs and solvent accessibilities (indicated by shaded boxes with black for buried and white for exposed positions) are shown in rows 3 and 4, respectively. As can be seen, fucose binding site(s), majority of which corresponds to the trisaccharide binding site in the resolved structure (PDB ID: 2obs), are located within variable, largely hydrophilic loops.
Figure 2.
Analysis of docking models for the complex of fucose and norovirus capsid protein. Right panel shows alternative fucose poses superimposed together. The corresponding interaction interfaces are shown in the left upper panel, with residues in contact with the ligand indicated by magenta. In the lower left panel, amino acid residues are aligned with the corresponding profiles: evolutionary conservation, physico-chemical properties, SS and RSA states.
In summary, POLYVIEW-MM provides a versatile Jmol-based interactive view of molecular trajectories and multiple conformers that can be generated by variety of simulation and modeling techniques. In addition, high-quality movies and static pictures can be generated to complement the interactive assessment of MM. These 3D representations are supplemented by customizable, intuitive 2D plots that capture at a glance the essence of conformational changes or differences between individual models to be assessed. POLYVIEW-MM combines visualization with structural and functional annotation by automatically mapping functional hot spots and structural features into analyzed models. The corresponding web server is publicly available, and it utilizes the same communication protocol and data submission/processing/retrieval technology as previously developed for POLYVIEW-2D (30) and POLYVIEW-3D (5). These servers are being widely used, logging more than 100 000 submissions from over 80 countries to date. Therefore, POLYVIEW-MM is expected to provide a fast and robust execution while processing significant number of requests from the users.
FUNDING
National Institutes of Health R01GM067823, R01A1055649; National Science Foundation GOALI:081163; Next Generation Biomedical Investigator Award (to A.P.) by the Center for Environmental Genetics funded by National Institute of Environmental Health Sciences (P30ES006096). Funding for open access charge: Grants from National Institutes of Health; Personal faculty member departmental accounts.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We would like to thank Roman Petrenko and Jacek Biesiada for their help with generating examples of MD and docking simulations. The authors also acknowledge the support of the Cincinnati Children’s Hospital Medical Center (CCHMC) and University of Cincinnati Medical College. We would like to dedicate this work to the author of PyMol, Warren DeLano, who passed away recently.
REFERENCES
- 1.Humphrey W, Dalke A, Schulten K. VMD: visual molecular dynamics. J. Mol. Graph. 1996;14:27–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 2.Guex N, Peitsch MC. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis. 1997;18:2714–2723. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- 3.DeLano WL. The PyMOL Molecular Graphics System. Palo Alto, CA: DeLano Scientific LLC; 2008. [Google Scholar]
- 4.Bohne-Lang A, Groch WD, Ranzinger R. AISMIG—an interactive server-side molecule image generator. Nucleic Acids Res. 2005;33:W705–709. doi: 10.1093/nar/gki438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Porollo A, Meller J. Versatile annotation and publication quality visualization of protein complexes using POLYVIEW-3D. BMC bioinformatics. 2007;8:316. doi: 10.1186/1471-2105-8-316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Autin L, Tuffery P. PMG: online generation of high-quality molecular pictures and storyboarded animations. Nucleic Acids Res. 2007;35:W483–W488. doi: 10.1093/nar/gkm277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zheng W, Brooks BR, Thirumalai D. Low-frequency normal modes that describe allosteric transitions in biological nanomachines are robust to sequence variations. Proc. Natl Acad. Sci. USA. 2006;103:7664–7669. doi: 10.1073/pnas.0510426103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gerstein M, Krebs W. A database of macromolecular motions. Nucleic Acids Res. 1998;26:4280–4290. doi: 10.1093/nar/26.18.4280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Phillips JC, Braun R, Wang W, Gumbart J, Tajkhorshid E, Villa E, Chipot C, Skeel RD, Kale L, Schulten K. Scalable molecular dynamics with NAMD. J. Comput. Chem. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brooks BR, Bruccoleri RE, Olafson BD, States DJ, Swaminathan S, Karplus M. CHARMM—a program for macromolecular energy, minimization, and dynamics calculations. J. Comput. Chem. 1983;4:187–217. [Google Scholar]
- 11.Lindahl E, Hess B, van der Spoel D. GROMACS 3.0: a package for molecular simulation and trajectory analysis. J. Mol. Model. 2001;7:306–317. [Google Scholar]
- 12.Goodsell DS, Morris GM, Olson AJ. Automated docking of flexible ligands: applications of AutoDock. J. Mol. recognit. 1996;9:1–5. doi: 10.1002/(sici)1099-1352(199601)9:1<1::aid-jmr241>3.0.co;2-6. [DOI] [PubMed] [Google Scholar]
- 13.Finn RD, Tate J, Mistry J, Coggill PC, Sammut SJ, Hotz HR, Ceric G, Forslund K, Eddy SR, Sonnhammer EL, et al. The Pfam protein families database. Nucleic Acids Res. 2008;36:D281–D288. doi: 10.1093/nar/gkm960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tusnady GE, Dosztanyi Z, Simon I. PDB_TM: selection and membrane localization of transmembrane proteins in the protein data bank. Nucleic Acids Res. 2005;33:D275–D278. doi: 10.1093/nar/gki002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bernstein FC, Koetzle TF, Williams GJ, Meyer EF, Jr, Brice MD, Rodgers JR, Kennard O, Shimanouchi T, Tasumi M. The Protein Data Bank: a computer-based archival file for macromolecular structures. J. Mol. Biol. 1977;112:535–542. doi: 10.1016/s0022-2836(77)80200-3. [DOI] [PubMed] [Google Scholar]
- 16.Dundas J, Ouyang Z, Tseng J, Binkowski A, Turpaz Y, Liang J. CASTp: computed atlas of surface topography of proteins with structural and topographical mapping of functionally annotated residues. Nucleic Acids Res. 2006;34:W116–W118. doi: 10.1093/nar/gkl282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Porollo A, Meller J. Prediction-based fingerprints of protein-protein interactions. Proteins. 2007;66:630–645. doi: 10.1002/prot.21248. [DOI] [PubMed] [Google Scholar]
- 18.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kabsch W, Sander C. Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers. 1983;22:2577–2637. doi: 10.1002/bip.360221211. [DOI] [PubMed] [Google Scholar]
- 20.Chothia C. The nature of the accessible and buried surfaces in proteins. J. Mol. Biol. 1976;105:1–12. doi: 10.1016/0022-2836(76)90191-1. [DOI] [PubMed] [Google Scholar]
- 21.Jones S, Thornton JM. Analysis of protein-protein interaction sites using surface patches. J. Mol. Biol. 1997;272:121–132. doi: 10.1006/jmbi.1997.1234. [DOI] [PubMed] [Google Scholar]
- 22.Chakrabarti P, Janin J. Dissecting protein-protein recognition sites. Proteins. 2002;47:334–343. doi: 10.1002/prot.10085. [DOI] [PubMed] [Google Scholar]
- 23.Martz E. Protein Explorer: easy yet powerful macromolecular visualization. Trends Biochem. Sci. 2002;27:107–109. doi: 10.1016/s0968-0004(01)02008-4. [DOI] [PubMed] [Google Scholar]
- 24.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 25.Nairn KM, Lyons RE, Mulder RJ, Mudie ST, Cookson DJ, Lesieur E, Kim M, Lau D, Scholes FH, Elvin CM. A synthetic resilin is largely unstructured. Biophys. J. 2008;95:3358–3365. doi: 10.1529/biophysj.107.119107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Elvin CM, Carr AG, Huson MG, Maxwell JM, Pearson RD, Vuocolo T, Liyou NE, Wong DC, Merritt DJ, Dixon NE. Synthesis and properties of crosslinked recombinant pro-resilin. Nature. 2005;437:999–1002. doi: 10.1038/nature04085. [DOI] [PubMed] [Google Scholar]
- 27.Petrenko R, Dickerson M, Naik R, Patnaik S, Beck T, Meller J. In: Proceedings of the 2009 International Conference on Bioinformatics & Computational Biology, BIOCOMP 2009. Arabnia HR, Yang MQ, editors. Vol. II. CSREA Press; 2009. pp. 598–603. [Google Scholar]
- 28.Tan M, Huang P, Meller J, Zhong W, Farkas T, Jiang X. Mutations within the P2 domain of norovirus capsid affect binding to human histo-blood group antigens: evidence for a binding pocket. J. Virol. 2003;77:12562–12571. doi: 10.1128/JVI.77.23.12562-12571.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cao S, Lou Z, Tan M, Chen Y, Liu Y, Zhang Z, Zhang XC, Jiang X, Li X, Rao Z. Structural basis for the recognition of blood group trisaccharides by norovirus. J. Virol. 2007;81:5949–5957. doi: 10.1128/JVI.00219-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Porollo AA, Adamczak R, Meller J. POLYVIEW: a flexible visualization tool for structural and functional annotations of proteins. Bioinformatics. 2004;20:2460–2462. doi: 10.1093/bioinformatics/bth248. [DOI] [PubMed] [Google Scholar]