Abstract
High-throughput gene-expression studies result in lists of differentially expressed genes. Most current meta-analyses of these gene lists include searching for significant membership of the translated proteins in various signaling pathways. However, such membership enrichment algorithms do not provide insight into which pathways caused the genes to be differentially expressed in the first place. Here, we present an intuitive approach for discovering upstream signaling pathways responsible for regulating these differentially expressed genes. We identify consistently regulated signature genes specific for signal transduction pathways from a panel of single-pathway perturbation experiments. An algorithm that detects overrepresentation of these signature genes in a gene group of interest is used to infer the signaling pathway responsible for regulation. We expose our novel resource and algorithm through a web server called SPEED: Signaling Pathway Enrichment using Experimental Data sets. SPEED can be freely accessed at http://speed.sys-bio.net/.
INTRODUCTION
Signal transduction pathways integrate information about the cellular environment and regulate the activity of a multitude of transcription factors, thereby controlling cellular processes, such as migration, proliferation and differentiation through context-dependent gene expression. While the techniques to measure transcript levels on a genome scale have matured and become affordable, it is not yet possible to get a global picture of the activity of signaling pathways in a high-throughput manner.
Typical strategies to analyze gene-expression profiles include searches for overrepresented biological functions, molecular mechanisms or pathways that the regulated genes are involved in, often utilizing annotation databases like Gene Ontology (1) and pathway databases such as KEGG (2). While these strategies have proven very useful to systematically organize the results, it is often forgotten that these analyses mainly detect pathways that are regulated in response to the perturbation—and not the cause of the observed regulatory pattern. If, for example, the MAPK signaling pathway has been found overrepresented in the regulated genes it only shows that the pathway is regulated in response to a perturbation, but not that the MAPK signaling pathway caused the gene regulation. Although genes regulated by a signaling pathway may involve genes that encode for members of the same pathway (feedback regulators), they may also regulate genes that are members of other pathways (transcriptional cross-talk) (3). Thus, pathway membership of regulated genes is unlikely to unveil the regulating pathway, and other strategies have to be employed in order to probe the origin of a certain gene-expression pattern.
One such computational strategy that is often used is the search of overrepresented binding sites in the promoter region of the regulated genes. This strategy is indeed suitable for finding the transcription factors causing the regulation. However, the method is seriously hampered by the poor specificity of computational prediction of transcription factor binding sites, due in large part to the high degeneracy and low information content of the recognized binding sequences (4). In the future, development in experimental strategies such as ChIP-chip or ChIP-seq will likely improve this situation. Still, links between signaling pathways and the transcription factors are only known for a limited number of cases. Thus, even if one has discovered which transcription factor is causing the regulation, in most cases one still cannot pin down the signaling pathway upstream of this transcription factor.
In order to infer signaling pathways that caused the regulation of a group of genes, we developed Signaling Pathway Enrichment using Experimental Data sets (SPEED), a data collection and algorithm that allows for identifying signaling pathways that cause an observed regulatory pattern. The key idea behind SPEED is that the same signaling pathway typically regulates a similar (small) core set of genes in most cell types, while different signaling pathways typically regulate different core sets of genes. We term such a set of consistently regulated genes ‘signature genes’ for that particular pathway. A list of regulated genes in an experiment can then be compared to a catalog of signature genes for different signaling pathways. In the following section, we present the SPEED web server with application to the analysis of two acute myeloid leukemia (AML) data sets.
MATERIALS AND METHODS
Selection and preprocessing of gene-expression data
Gene-expression data sets were identified manually in the gene-expression omnibus (GEO) database based on their abstracts. We concentrated our search on single-perturbation experiments measured within 4 h after treatment to limit secondary effects. This time span represents the shortest interval, which still allowed collection of a sufficient number of data sets in most pathways. If, however, not enough such data sets were available for a particular pathway, long-term experiments such as stable overexpression or knockdown experiments were included if their effect on signature genes was assumed to be prominent.
Custom R-scripts were written to download and process the selected expression data sets automatically. Data for each micro-array experiment within the data set were subjected to quantile normalization and log-transformation if needed. Finally, a LOESS model for the standard deviation as a function of expression value was trained either on replicate experiments (when available) or on all experiments and was used to compute Z-scores. Z-score (x) = (x − μ)/σ, where μ is the sample mean and σ is the standard deviation. The probes were annotated with Entrez Gene identifiers using GEOquery.
Implementation
SPEED runs on an Apache server with the pre-processed micro-array data stored in an SQLite 3.0 database (see Supplementary Figure S2 for database schema). The SPEED algorithm is implemented in Python 2.5 with dependency on the scipy 0.7.0 (http://www.scipy.org/) package. Graphical output is dynamically generated using the Google Chart API (http://code.google.com/apis/chart/).
Clustering
Hierarchical clustering was performed using the statistical package R, clusters were linked using Ward’s method. The distance between two pathways was defined as: −log [# common signature genes/min (# signature genes for each set)].
Validation using literature gene lists
Gene lists were extracted directly from the manuscripts or supplementary data and were not further processed (see Supplementary Table S1 for references). Raw data were not available for these lists. Every significant hit in SPEED had a false discovery rate (FDR) < 0.05.
FANTOM4 data processing
The FANTOM4 data set was downloaded as tab delimited text files from the Center for Information Biology gene EXpression (CIBEX) database (http://cibex.nig.ac.jp/cibex2/ExperimentMiame.do?queryExperimentalDesignAccession=CBX47). Each of the 52 transcription factor knockdowns in THP-1 cell line were conducted in three replicates. Probe expression values were divided by appropriate controls and the ratios were log (base 2) transformed. Probes were only considered valid if the transcript was expressed in all three replicates for at least half of the 52 transcription factor knockdowns. Valid probe IDs were converted to Entrez Gene IDs and used as background. Genes with at least 2-fold differential median expression were considered for input to SPEED.
RESULTS
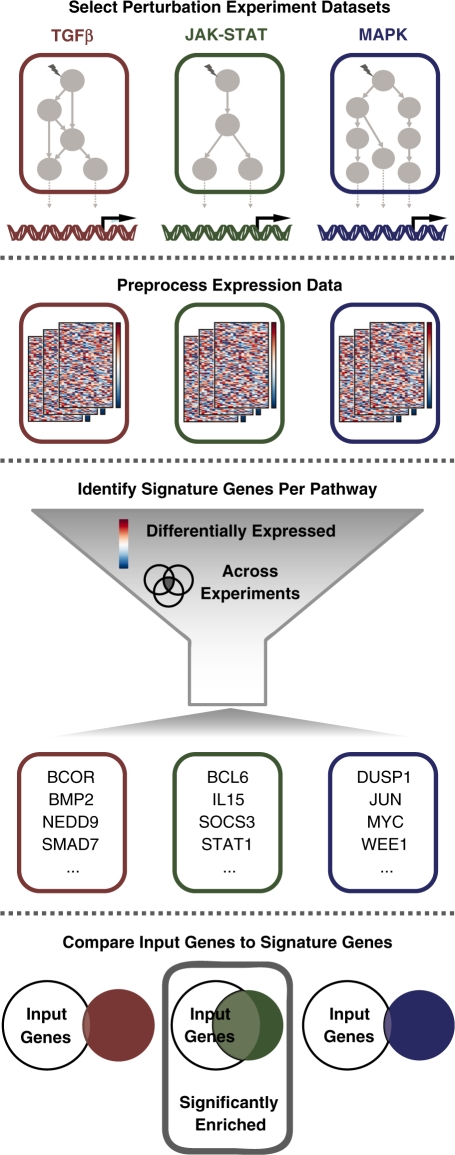
We define signature genes as genes that are consistently regulated by a particular pathway across many experiments. Overrepresentation of such signature genes in a list of differentially expressed genes can then hint at the signaling pathway that caused the regulation. We set up a pipeline to compile publicly available expression data sets from pathway perturbation experiments, store them into a database, extract signature genes, and finally detect significant overlap with a user-supplied gene group via a web server (http://speed.sys-bio.net/, see schematics in Figure 1).
Figure 1.
Overview of the SPEED algorithm. The SPEED algorithm is based on the identification of signature genes that are consistently regulated by specific signaling pathways using publicly available micro-array data. Gene-expression data sets from single-pathway perturbation experiments were manually selected from the GEO database. Next, gene-expression values from the selected database were automatically processed using custom R-scripts, and expression changes were stored as Z-score rank percentiles in the SPEED database. The SPEED web server extracts signature genes per pathway on the fly based on user-specified parameters describing the level of differential expression (Z-score percentile; ex: top 1%) and the level of consistency across experiments (percentage of experimental data sets where a gene is differentially expressed; ex: at least 20%). Users can compare their own gene sets against the extracted signature genes to identify modulated upstream signaling pathways.
Compilation of a database to define signature genes for signaling pathways
In order to extract signature genes, we selected a set of 11 signaling pathways: TGF-β, H2O2, TLR, IL-1, MAPK, PI3K, MAPK+PI3K, Wnt, JAK-STAT, TNF-α and VEGF. For each signaling pathway, we manually searched the GEO database (5) for gene-expression data sets where this signaling pathway is specifically perturbed (see ‘Materials and Methods’ for selection criteria). The data sets were automatically downloaded from GEO (6), annotated and normalized. The expression changes per probe were then transformed into Z-scores per gene (see ‘Materials and Methods’ section for details). Subsequently, the gene ID, expression rank and Z-score rank were stored in the SPEED database. Currently, 215 sets of micro-array experiments are stored in the database, resulting in almost 6 million expression values for 21 485 human genes.
From this database, signature genes for each pathway can be extracted. We define three criteria for a signature gene: it has to be (i) expressed and (ii) regulated (iii) across several experiments for each pathway. As the stored micro-array data originates from different sources and even platforms, we chose to apply thresholds based on percentiles to select the signature genes. For each of the data sets, genes are defined as expressed if their expression value rank is higher than a pre-determined threshold [criteria (i); default: top 50%). Next, regulated genes are extracted based on the rank of their Z-score [criteria (ii); default: top 1%). Finally, genes are selected as signature genes if they are expressed and regulated in more than a pre-determined percentage of data sets for a pathway [criteria (iii); default: >20%). Users can adjust these parameters to create lists of signature genes on the fly.
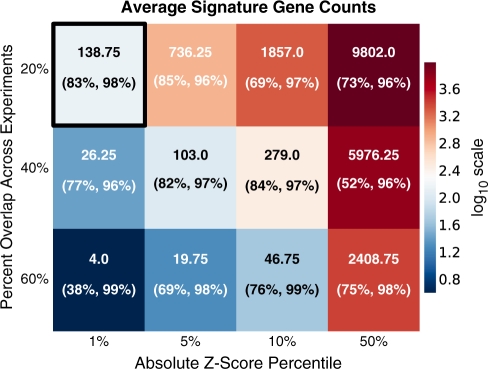
The mean number of signature genes per pathway using the default parameters is 139. The signature gene lists for any given parameter set can be downloaded as tab delimited text directly from the SPEED web server. Figure 2 lists the mean number of signature genes per pathway for various parameters.
Figure 2.
Average number of SPEED signature genes. SPEED signature genes are sensitive to user-specified parameters. Average signature genes per pathway without the uniqueness constraint are listed as a function of Z-score percentile and percent overlap across experiments. The heat map corresponds to log10 of the average number of signature genes. The sensitivity and specificity is noted in parenthesis. The values for the default parameter set of Z-score percentile ≤1% and percent overlap ≥20% are boxed. Our choice of default parameter set, as well as recommended parameters (in black text) are determined ad hoc based on their biological meaning, resulting number of signature genes and performance metrics. Only the default pathways are considered here and bottom 50% expressed genes are discarded for all calculations.
Overlap of signature genes across pathways
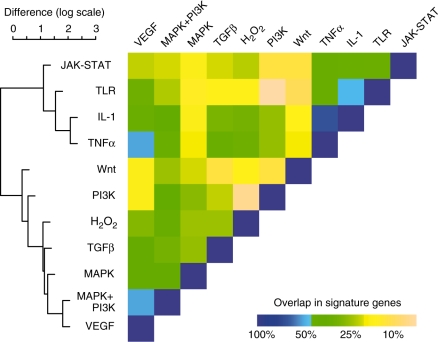
Since signaling pathways form highly interconnected networks, the definition of distinct signaling pathways is difficult and often arbitrary. We therefore analyzed the overlap between signature genes between all pairs of pathways and noticed that there is considerable overlap. In order to group similar pathways, we performed hierarchical cluster analysis (Figure 3). Clustering revealed that the major clusters separate pathways involved in immune response from pathways controlling cell cycle and cell growth. Furthermore, VEGF signaling seems very closely related to pathways from both clusters (MAPK+PI3K signaling and TNF-α signaling). Also, signaling triggered by IL-1 and TNF-α shows strong overlap in the signature genes, probably due to the shared downstream IKK-β/NF-κB signaling cascade (7). We therefore added a feature in SPEED to select only those signature genes that are unique to each pathway. We note, however, that under such a uniqueness constraint the signature genes depend strongly on the thresholds chosen, and the predictive power of the algorithm does not improve (Figure 2; Supplementary Figure S1). Additionally, since we expect a strong crosstalk between pathways, non-unique signature genes seem to be a more biologically reasonable choice. We also decided to give the user the possibility to restrict their analysis on a subset of pathways. The default setting restricts the analysis on four major pathways JAK-STAT, MAPK+PI3K, TGF-β and TLR. While users have complete control in their choice of parameters, the selected parameters should be biologically reasonable (Figure 2 describes our recommended parameters).
Figure 3.
Signaling pathway crosstalk demonstrated by overlapping signature genes. The percent overlap between signature genes between all pairs of pathways is displayed as a heat map with higher overlap suggesting greater crosstalk between the respective signaling pathways. Pathway similarity is calculated as the negative log of the percent overlap and similar pathways are grouped using hierarchical clustering. Two major clusters are realized, separating pathways involved in immune response (JAK-STAT, TLR, IL-1 and TNF-α) from pathways controlling cell cycle.
Detecting overrepresented signature genes
In order to detect overrepresented signature genes, we apply Fisher’s exact test with a multiple hypothesis correction. For each pathway, it is tested whether the corresponding signature genes are overrepresented in the user-supplied list. A P-value is provided to indicate the significance for the overrepresentation. To adjust for multiple hypotheses, an FDR for the P-value (8) is also provided. Users can restrict the analysis to a subset of genes in the database by submitting a list of background genes. For example, if one submits a list of differentially expressed genes from a micro-array study, we would recommend using all significantly expressed genes from that micro-array study as background.
Validation
In order to test how well SPEED can predict modulated signaling pathway activity, we collected gene lists from literature with known signaling pathway perturbation. Note that gene-expression data from these literature sources was not integrated into SPEED. A result was considered to be true positive if the top-ranked pathway prediction matched the actual perturbed pathway and the FDR was below 0.05. Table 1 describes the validation results for the default pathways (see Supplementary Table S1 for details). The overall sensitivity, calculated as the fraction of all literature sources with correct pathway predictions, using default parameters was found to be 78%. Since traditional signaling pathway membership analyses do not aim at predicting causal pathways, SPEED outperforms GATHER (9), which correctly predicts the upstream pathway with a sensitivity of 29%.
Table 1.
Validation of the SPEED algorithm on gene lists from independent literature sources
| Pathway | Test sets | Correct top-ranking events (sensitivity) |
Significantly overrepresented events (sensitivity) |
||
|---|---|---|---|---|---|
| SPEED (%) | GATHER (%) | SPEED (%) | GATHER (%) | ||
| JAK-STAT | 10 | 8 (80) | 1 (10) | 10 (100) | 7 (70) |
| TGF-β | 5 | 4 (80) | 2 (40) | 5 (100) | 2 (40) |
| MAPK+PI3K | 6 | 4 (67) | 1 (17) | 6 (100) | 6 (100) |
| TLR | 6 | 5 (83) | 3 (50) | 6 (100) | 3 (50) |
| Total | 27 | 21 (78) | 7 (29) | 27 (100) | 18 (65) |
SPEED results from 27 literature-derived gene lists for the four default pathways are summarized. As compared to traditional signaling pathway membership analysis using GATHER, SPEED correctly predicts the perturbed signaling pathway as the top-ranking result at a higher rate (78% compared to 29%). SPEED also outperforms GATHER in identifying the correct pathway as one of the significantly overrepresented pathways (FDR ≤ 0.05 for SPEED and no threshold for GATHER).
Due to limited number of available positive sets in literature, we also conducted leave-one-out cross validation to get a better estimate of sensitivity. Once again, we considered a result to be true positive if the top-ranking predicted pathway was indeed the one that was perturbed. Using default parameter values, the sensitivity was calculated to be 83% of 130 tests. The classification problem here is not binary and therefore the expected sensitivity for random predictions is far <50%. In order to estimate the specificity, the fraction of negative input sets correctly identified as negative, we ran SPEED on 200 negative sets comprised of randomly selected genes from the database. The size of the negative sets was randomly chosen between 50 and 200 genes. The overall specificity and precision (the proportion of true positive results against all positive results) was calculated to be 98 and 96%, respectively. Figure 2 notes the sensitivity and specificity for different parameters.
Comparison with pathway membership databases
Validation using the literature derived gene sets indicated a difference between genes regulated by a pathway and gene products that are members of the pathway. To verify the disparity between regulated genes and pathway members, we searched regulated genes from each SPEED experimental data set for overrepresentation of acting pathway members in KEGG Pathway (2), BioCarta and Panther (10) databases. 39% of all data sets had significant enrichment of the acting pathway members as determined by KEGG Pathway. 9 and 24% of all data sets had significant enrichment of the corresponding pathway in BioCarta and Panther, respectively. These findings suggest some degree of transcriptional feedback where genes regulated by a pathway translate to protein members within the same pathway. However, only 4 and 7% of all data sets had the acting pathway ranked as the top one in KEGG and Panther, respectively. No experimental data set had the acting pathway ranked as the top one in BioCarta (see Supplementary Tables S4 and S5 and Figures 3 and 4, for detailed results). The low top-rank percentage reasserts the unfavorable use of pathway membership databases for identifying pathways regulating a list of genes. Consequently, the SPEED data sets and signature genes are fundamentally different than lists of pathway members found in existing membership databases.
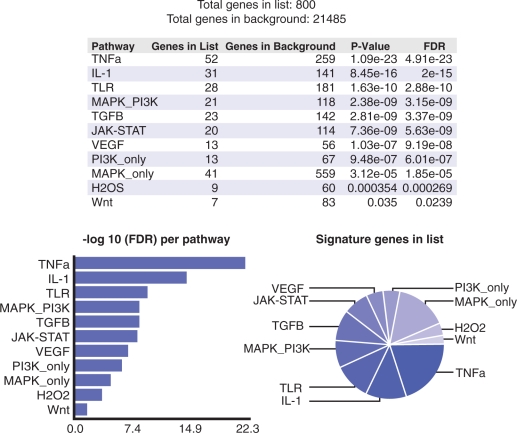
Figure 4.
SPEED analysis of downregulated genes in high-risk AML patients with CEBPa mutations. Screenshot of SPEED results shows TNF-α as the top-ranking upstream signaling pathway for target genes downregulated in high-risk AML patients with CEBPa mutations versus CEBPa wild-type. The pathways are sorted by FDR and graphical outputs aid in interpretation of the results.
Web server
The scope of the SPEED web server is the identification of causal signaling pathways given a list of input human genes. Thus, we implemented the web server with an unencumbered user interface where the user can input gene lists in widely adopted formats including Entrez Gene ID, gene symbol, Uniprot, GI number, Refseq, Ensembl and IPI. Any other gene identifiers can be easily converted to those formats using available conversion tools such as Clone ID converter (11), DAVID bioinformatics resources (12) or the UniProt ID mapping service (13). Upon submission, the user is directed to the output page where the results are presented as a list of signaling pathways ordered by their significance with number of signature genes present in the input, P-values and FDRs for each pathway. Graphical outputs, such as a pie chart representing the distribution of signature genes in the input list and a bar chart describing the relative FDRs, further aid in interpretation of the results (Figure 4). The symbols of the signature genes present in the input are provided with hyperlinks to the respective NCBI Entrez Gene web page. Separate web pages are provided for database access to retrieve signature genes as tab delimited text or to browse through experimental details such as cell type, perturbation strategy and length of perturbation with hyperlinks to the original gene-expression data sources.
In addition to the web interface, we provide download options for the raw data as tab delimited text or as an SQLite database, which is a single cross-platform disk file, together with Python source code to facilitate local programmatic access to SPEED functionality. All SPEED code is open-source and free to use.
In the following sections, we demonstrate the functionality of SPEED through analysis of two AML data sets.
Identification of signaling pathways downregulating genes in high-risk AML patients with CEBPa mutations
Patients with AML harbor mutations in one or more genes such as FLT3, NPM1, c-KIT and CEBPa (14). Some mutations like internal tandem duplication in FLT3 (FLT3-ITD) are associated with high-risk AML patients and confer poor outcomes (14). Marcucci et al. (14) conducted a study to search for prognostic markers in cytogenetically normal AML patients with high-risk molecular features (FLT3-ITD and/or NPM1 wild-type) with or without CEBPa mutations. The study identified 928 genes that were downregulated in CEBPa mutants as compared to wild-type. We searched the downregulated genes for enriched pathway annotations using GATHER (9) and DAVID (12), using standard parameters (Bayes factor ≥6 and corrected P-value ≤ 0.05 for GATHER and DAVID, respectively). No pathways were identified as containing a significant number of members in the input gene list, which suggests that the downregulated genes have little effect on signaling pathways.
As previously noted, searching for membership enrichment does not provide insight into the signaling pathways that causally regulate a list of genes. We searched the same list of downregulated genes using SPEED (default parameters with all pathways selected) and identified TNF-α as the top-ranking pathway (Figure 4). Interestingly, the top three pathways, TNF-α, IL-1 and TLR, form a single cluster (Figure 3).
Predicting signaling pathways upstream of 24 transcription factors in THP-1 cells
In the recently published FANTOM4 study, the consortium knocked down 52 transcription factors in the AML cell line THP-1 (15) and measured changes in the transcriptome. We reasoned that if the target genes of the transcription factors overlap significantly with signature genes of a signaling pathway, the pathway is likely to be upstream of that transcription factor. We therefore used this data to see whether SPEED can be used to systematically infer relationships between signaling pathways, transcription factors and regulated genes. We extracted 52 lists of differentially expressed genes corresponding to each transcription factor knockdown (see ‘Materials and Methods’ section for details) and ran SPEED to identify upstream signaling pathways. SPEED identified at least one signaling pathway upstream of 24 transcription factors. Table 2 describes the top-ranking signaling pathway for each transcription factor (see Supplementary Tables S2 and S3 for all predictions). While for some of the factors, the identified pathway is known to be upstream (indicated in the table), for others our results are predictions that may provide starting points for experiments.
Table 2.
Identification of signaling pathways upstream of transcription factor in THP-1 cells
| Top-ranked pathway | Transcription factor |
|---|---|
| MAPK+PI3K | CEBPA (27), EGR1, (28), ETS2 (29), FLI1 (30), MLLT3, MYBL2 (31), NFE2L1, NFYA, NRASa, SP1 (32), ZNF238 |
| JAK-STAT | FOXJ3, FOXP1, GFI1 (33), MXI1, RUNX1 (34), STAT1 (35), YY1 |
| TLR | BMI1, CEBPG, GATA2 (36), IRF8 (37), SPI1 (38) |
| TGF-β | MYB (39) |
SPEED was run programatically on lists of differentially expressed genes following 52 transcription factor knockdowns. For 24 transcription factors, the corresponding differentially expressed gene lists had an overrepresentation of at least one upstream signaling pathway. The top-ranking signaling pathway with a minimum FDR of 0.05 is listed for each of the 24 transcription factors. Literature references for transcription factors known to be associated with identified signaling pathways are noted.
DISCUSSION
The analysis of gene lists obtained from high-throughput experiments is a classical problem in bioinformatics. Most approaches, however, search for overrepresented functions, processes or pathways in the regulated gene group and are thus only suitable to identify the affected signaling pathways and not the pathways that caused the observed changes in gene expression. With SPEED, we focus on targets of signaling pathways by automatically deriving signature genes, i.e. genes that are typically regulated in a variety of cell types when the activity of a specific pathway is altered. While existing web servers also compare user gene lists with disease related or single-experiment signatures derived from micro-array data (16–21), SPEED, to our knowledge, is the only web server that integrates heterogeneous micro-array data for the purpose of identifying causal signaling pathway relationships. Additionally, SPEED signature gene lists can be readily incorporated into established gene list comparison tools like Gene Set Enrichment Analysis (22).
The number of pathways in SPEED is currently limited by publicly available gene-expression data from pathway perturbation experiments. Currently, SPEED contains 11 signaling pathways. However, the presence of pathway perturbation data in GEO suggests that these 11 pathways are of special interest to researchers and are thus the major signaling pathways responsible for many phenotypes studied using gene-expression technologies. Nevertheless, we expect that the number of SPEED pathways will increase in the future as microarrays continue to become more affordable. Furthermore, we can readily incorporate quantitative next-generation sequencing data as the field matures. To facilitate user suggestions, we have included a form on the web server for recommending new pathways or data.
It is important to note that not all pathway perturbation data should be included in SPEED. The reliability and biological meaning of SPEED signatures crucially depends on the nature of the experiments. For example, we aimed at selecting experiments that measured gene expression at early time points in order to reduce any indirect or long-term effects. To further reduce indirect effects, we selected experiments such that single pathways were primarily perturbed. While some perturbations may affect more than a single pathway, the reproducibility over multiple experiments via the overlap parameter can account for noise in the data as long as the pathway is the primarily perturbed one. However, if pathways are strongly coupled or converge on the same transcription factors, such as the MAPK and PI3K/AKT pathways (23), it is necessary to combine them into a single pathway description. The SPEED pathway labeled MAPK+PI3K is an example of combining coupled pathways.
Validation using literature-derived gene lists indicated that SPEED performs well in predicting upstream causal pathway-perturbation events with a sensitivity of 78%. However, it is possible that other cellular processes also regulate the same genes as SPEED signaling pathways and are the actual causal influences. Since the predicted signaling pathways are derived based on correlations with gene signatures, results should be considered as a starting point for further investigation into the actual events leading to the regulated set of genes.
The identification of signaling pathways that cause observed changes in gene expression is important, since aberrant signaling pathways have been implicated in diseases such as diabetes (24) and several cancers (25,26). We exemplify the utility of SPEED by identifying upstream signaling pathways in two AML studies. For high-risk AML patients with and without CEBPa mutations, SPEED identified TNF-α signaling as the top candidate that caused the observed differences between the two groups of patients. Given that high-risk AML patients with CEBPa mutations have better clinical outcomes, SPEED results may be taken as a starting point to narrow down the search for the differences in outcome by focusing on differences in TNF-α signaling.
Using micro-array data from knockdown of transcription factors in an AML cell line, we aimed at identifying links between transcription factors and upstream signaling pathways. We searched for associations between signature genes of signaling pathways and the targets of the transcription factors. For 24 factors, we could establish such associations, of which some are already known, whereas others, like MAPK+PI3K to ZNF238, are novel predictions. Thus, SPEED facilitates completing the picture of cell signaling events leading to differential expression of genes via a layer of transcription factors.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Germany’s Federal Ministry for Education and Research (BMBF) (grant Forsys Partner to N.B.); the European Commission (grant number CancerSys HEALTH-F4-2008-223188 to N.B.); National Science Foundation Integrative Graduate Education and Research Traineeship (IGERT) (grant number DGE-0654108 to J.P.). Funding for open access charge: National Science Foundation Integrative Graduate Education and Research Traineeship (IGERT) (grant number DGE-0654108).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Rositsa Koleva and Eric D. Smith for helpful discussions.
REFERENCES
- 1.Harris MA, Clark J, Ireland A, Lomax J, Ashburner M, Foulger R, Eilbeck K, Lewis S, Marshall B, Mungall C, et al. The Gene Ontology (GO) database and informatics resource. Nucleic Acids Res. 2004;32:D258–D261. doi: 10.1093/nar/gkh036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kanehisa M, Goto S, Furumichi M, Tanabe M, Hirakawa M. KEGG for representation and analysis of molecular networks involving diseases and drugs. Nucleic Acids Res. 38:D355–D360. doi: 10.1093/nar/gkp896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Legewie S, Herzel H, Westerhoff HV, Bluthgen N. Recurrent design patterns in the feedback regulation of the mammalian signalling network. Mol. Syst. Biol. 2008;4:190. doi: 10.1038/msb.2008.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wasserman WW, Sandelin A. Applied bioinformatics for the identification of regulatory elements. Nat. Rev. Genet. 2004;5:276–287. doi: 10.1038/nrg1315. [DOI] [PubMed] [Google Scholar]
- 5.Barrett T, Edgar R. Mining microarray data at NCBI's Gene Expression Omnibus (GEO)*. Methods Mol. Biol. 2006;338:175–190. doi: 10.1385/1-59745-097-9:175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sean D, Meltzer PS. GEOquery: a bridge between the Gene Expression Omnibus (GEO) and BioConductor. Bioinformatics. 2007;23:1846–1847. doi: 10.1093/bioinformatics/btm254. [DOI] [PubMed] [Google Scholar]
- 7.Vallabhapurapu S, Karin M. Regulation and function of NF-kappaB transcription factors in the immune system. Annu. Rev. Immunol. 2009;27:693–733. doi: 10.1146/annurev.immunol.021908.132641. [DOI] [PubMed] [Google Scholar]
- 8.Bluthgen N, Kielbasa SM, Herzel H. Inferring combinatorial regulation of transcription in silico. Nucleic Acids Res. 2005;33:272–279. doi: 10.1093/nar/gki167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chang JT, Nevins JR. GATHER: a systems approach to interpreting genomic signatures. Bioinformatics. 2006;22:2926–2933. doi: 10.1093/bioinformatics/btl483. [DOI] [PubMed] [Google Scholar]
- 10.Thomas PD, Campbell MJ, Kejariwal A, Mi H, Karlak B, Daverman R, Diemer K, Muruganujan A, Narechania A. PANTHER: a library of protein families and subfamilies indexed by function. Genome Res. 2003;13:2129–2141. doi: 10.1101/gr.772403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Alibes A, Yankilevich P, Canada A, Diaz-Uriarte R. IDconverter and IDClight: conversion and annotation of gene and protein IDs. BMC Bioinformatics. 2007;8:9. doi: 10.1186/1471-2105-8-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 13.Consortium U. The Universal Protein Resource (UniProt) in 2010. Nucleic Acids Res. 2010;38:D142–D148. doi: 10.1093/nar/gkp846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Marcucci G, Maharry K, Radmacher MD, Mrozek K, Vukosavljevic T, Paschka P, Whitman SP, Langer C, Baldus CD, Liu CG, et al. Prognostic significance of, and gene and microRNA expression signatures associated with, CEBPA mutations in cytogenetically normal acute myeloid leukemia with high-risk molecular features: a Cancer and Leukemia Group B Study. J. Clin. Oncol. 2008;26:5078–5087. doi: 10.1200/JCO.2008.17.5554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Suzuki H, Forrest AR, van Nimwegen E, Daub CO, Balwierz PJ, Irvine KM, Lassmann T, Ravasi T, Hasegawa Y, de Hoon MJ, et al. The transcriptional network that controls growth arrest and differentiation in a human myeloid leukemia cell line. Nat. Genet. 2009;41:553–562. doi: 10.1038/ng.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Culhane AC, Schwarzl T, Sultana R, Picard KC, Picard SC, Lu TH, Franklin KR, French SJ, Papenhausen G, Correll M, et al. GeneSigDB–a curated database of gene expression signatures. Nucleic Acids Res. 38:D716–D725. doi: 10.1093/nar/gkp1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wu J, Qiu Q, Xie L, Fullerton J, Yu J, Shyr Y, George AL, Jr, Yi Y. Web-based interrogation of gene expression signatures using EXALT. BMC Bioinformatics. 2009;10:420. doi: 10.1186/1471-2105-10-420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang SD, Gant TW. A simple and robust method for connecting small-molecule drugs using gene-expression signatures. BMC Bioinformatics. 2008;9:258. doi: 10.1186/1471-2105-9-258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cahan P, Ahmad AM, Burke H, Fu S, Lai Y, Florea L, Dharker N, Kobrinski T, Kale P, McCaffrey TA. List of lists-annotated (LOLA): a database for annotation and comparison of published microarray gene lists. Gene. 2005;360:78–82. doi: 10.1016/j.gene.2005.07.008. [DOI] [PubMed] [Google Scholar]
- 20.Newman JC, Weiner AM. L2L: a simple tool for discovering the hidden significance in microarray expression data. Genome Biol. 2005;6:R81. doi: 10.1186/gb-2005-6-9-r81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rhodes DR, Kalyana-Sundaram S, Mahavisno V, Varambally R, Yu J, Briggs BB, Barrette TR, Anstet MJ, Kincead-Beal C, Kulkarni P, et al. Oncomine 3.0: genes, pathways, and networks in a collection of 18,000 cancer gene expression profiles. Neoplasia. 2007;9:166–180. doi: 10.1593/neo.07112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl Acad. Sci. USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tullai JW, Schaffer ME, Mullenbrock S, Kasif S, Cooper GM. Identification of transcription factor binding sites upstream of human genes regulated by the phosphatidylinositol 3-kinase and MEK/ERK signaling pathways. J. Biol. Chem. 2004;279:20167–20177. doi: 10.1074/jbc.M309260200. [DOI] [PubMed] [Google Scholar]
- 24.Jauregui A, Mintz DH, Mundel P, Fornoni A. Role of altered insulin signaling pathways in the pathogenesis of podocyte malfunction and microalbuminuria. Curr. Opin. Nephrol. Hypertens. 2009;18:539–545. doi: 10.1097/MNH.0b013e32832f7002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gottesman MM. Report of a meeting: molecular basis of cancer therapy. J. Natl Cancer Inst. 1994;86:1277–1285. doi: 10.1093/jnci/86.17.1277. [DOI] [PubMed] [Google Scholar]
- 26.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 27.Tomizawa M, Saisho H. Insulin-like growth factor (IGF)-II regulates CCAAT/enhancer binding protein alpha expression via phosphatidyl-inositol 3 kinase in human hepatoblastoma cell lines. J. Cell Biochem. 2007;102:161–170. doi: 10.1002/jcb.21293. [DOI] [PubMed] [Google Scholar]
- 28.Sgambato V, Pages C, Rogard M, Besson MJ, Caboche J. Extracellular signal-regulated kinase (ERK) controls immediate early gene induction on corticostriatal stimulation. J. Neurosci. 1998;18:8814–8825. doi: 10.1523/JNEUROSCI.18-21-08814.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang BS, Hauser CA, Henkel G, Colman MS, Van Beveren C, Stacey KJ, Hume DA, Maki RA, Ostrowski MC. Ras-mediated phosphorylation of a conserved threonine residue enhances the transactivation activities of c-Ets1 and c-Ets2. Mol Cell Biol. 1996;16:538–547. doi: 10.1128/mcb.16.2.538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jinnin M, Ihn H, Mimura Y, Asano Y, Yamane K, Tamaki K. Matrix metalloproteinase-1 up-regulation by hepatocyte growth factor in human dermal fibroblasts via ERK signaling pathway involves Ets1 and Fli1. Nucleic Acids Res. 2005;33:3540–3549. doi: 10.1093/nar/gki648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hanada N, Lo HW, Day CP, Pan Y, Nakajima Y, Hung MC. Co-regulation of B-Myb expression by E2F1 and EGF receptor. Mol. Carcinog. 2006;45:10–17. doi: 10.1002/mc.20147. [DOI] [PubMed] [Google Scholar]
- 32.Milanini-Mongiat J, Pouyssegur J, Pages G. Identification of two Sp1 phosphorylation sites for p42/p44 mitogen-activated protein kinases: their implication in vascular endothelial growth factor gene transcription. J. Biol. Chem. 2002;277:20631–20639. doi: 10.1074/jbc.M201753200. [DOI] [PubMed] [Google Scholar]
- 33.Rodel B, Tavassoli K, Karsunky H, Schmidt T, Bachmann M, Schaper F, Heinrich P, Shuai K, Elsasser HP, Moroy T. The zinc finger protein Gfi-1 can enhance STAT3 signaling by interacting with the STAT3 inhibitor PIAS3. EMBO J. 2000;19:5845–5855. doi: 10.1093/emboj/19.21.5845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sheng Z, Wang SZ, Green MR. Transcription and signalling pathways involved in BCR-ABL-mediated misregulation of 24p3 and 24p3R. EMBO J. 2009;28:866–876. doi: 10.1038/emboj.2009.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hu X, Ivashkiv LB. Cross-regulation of signaling pathways by interferon-gamma: implications for immune responses and autoimmune diseases. Immunity. 2009;31:539–550. doi: 10.1016/j.immuni.2009.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu W, Zhu ZQ, Wang W, Zu SY, Zhu GJ. Crucial roles of GATA-2 and SP1 in adrenomedullin-affected expression of tissue factor pathway inhibitor in human umbilical vein endothelial cells exposed to lipopolysaccharide. Thromb. Haemost. 2007;97:839–846. [PubMed] [Google Scholar]
- 37.Laricchia-Robbio L, Tamura T, Karpova T, Sprague BL, McNally JG, Ozato K. Partner-regulated interaction of IFN regulatory factor 8 with chromatin visualized in live macrophages. Proc. Natl Acad. Sci. USA. 2005;102:14368–14373. doi: 10.1073/pnas.0504014102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ishii J, Kitazawa R, Mori K, McHugh KP, Morii E, Kondo T, Kitazawa S. Lipopolysaccharide suppresses RANK gene expression in macrophages by down-regulating PU.1 and MITF. J. Cell Biochem. 2008;105:896–904. doi: 10.1002/jcb.21886. [DOI] [PubMed] [Google Scholar]
- 39.Classen S, Zander T, Eggle D, Chemnitz JM, Brors B, Buchmann I, Popov A, Beyer M, Eils R, Debey S, et al. Human resting CD4+ T cells are constitutively inhibited by TGF beta under steady-state conditions. J. Immunol. 2007;178:6931–6940. doi: 10.4049/jimmunol.178.11.6931. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.