Abstract
SPOT (http://spot.cgsmd.isi.edu), the SNP prioritization online tool, is a web site for integrating biological databases into the prioritization of single nucleotide polymorphisms (SNPs) for further study after a genome-wide association study (GWAS). Typically, the next step after a GWAS is to genotype the top signals in an independent replication sample. Investigators will often incorporate information from biological databases so that biologically relevant SNPs, such as those in genes related to the phenotype or with potentially non-neutral effects on gene expression such as a splice sites, are given higher priority. We recently introduced the genomic information network (GIN) method for systematically implementing this kind of strategy. The SPOT web site allows users to upload a list of SNPs and GWAS P-values and returns a prioritized list of SNPs using the GIN method. Users can specify candidate genes or genomic regions with custom levels of prioritization. The results can be downloaded or viewed in the browser where users can interactively explore the details of each SNP, including graphical representations of the GIN method. For investigators interested in incorporating biological databases into a post-GWAS SNP selection strategy, the SPOT web tool is an easily implemented and flexible solution.
INTRODUCTION
Due to corrections for multiple testing and limited sample sizes, genome-wide association studies (GWAS) often lack the statistical power to discover statistically significant associations between phenotype and genotype (1). Therefore when a single nucleotide polymorphism (SNP) shows relatively strong evidence of genetic association, that is, is among the top signals from the study, the next step is to genotype the SNP in additional independent samples in order to prove the association is not simply due to chance. The strategy for selecting SNPs for additional genotyping could be simple, such as ranking the SNPs by their P-values from statistical tests for association, or somewhat complex if certain biological considerations are taken into account (2). Once a set of SNPs has been confirmed to be associated with the phenotype, the next logical steps include functional experiments that attempt to isolate the precise molecular genetic mechanism, such as the effect of the genetic variant on transcription, which may act by a direct modification to the amino acid sequence or structure of the protein product, or by an effect on a regulatory mechanism. Functional experiments can be very costly, raising the question of how to prioritize SNPs to maximize returns.
Even when there is only a single confirmed SNP, linkage disequilibrium (LD) with other SNPs can make it very difficult to isolate the true causal polymorphism. When a single SNP is confirmed to be associated with a phenotype and is in strong LD with several other SNPs, the genotypes at these so-called ‘LD proxies’ will be very similar. While this property is often exploited by manufacturers of commercial SNP microarrays (3) to reduce the number of SNPs required for genotyping, in this case it proves to be a serious problem because all the LD proxies show the same evidence for association and no amount of further genotyping will resolve this ambiguity. Instead, expensive and time-consuming functional experiments, possibly involving model organisms, must be conducted in an effort to identify the true causal variant.
In these scenarios, the known biological properties of the variants can prove to be useful in formulating a prioritization strategy. When selecting SNPs for further study after a GWAS, such as genotyping in a replication sample, investigators may choose to prioritize solely based on the statistical evidence for genotype–phenotype correlation; in other words use a single P-value threshold. If there are sufficient resources to select all SNPs above a maximum desired P-value, which could be determined by the combination of a minimum desired effect size and specification of statistical power under some reasonable transmission models, then prioritization by P-value alone is logical. When resources are limited, certain genes and genomic regions may be given higher priority, such as selecting all SNPs with P < 10−4 and SNPs in certain genes with P < 10−2. Because these ranges of P-values may involve many false positives, care should be used in determining the thresholds, and a careful evaluation of statistical power should be used to determine the goals of the study, such as specifying a desired minimum effect size. When the evidence for association boils down to a single SNP and all its LD proxies, the P-values are all nearly identical and so biological prioritization becomes particularly attractive. This kind of prioritization strategy allows investigators to maximize the return on resources while implementing their specific biological priorities.
We recently introduced the genomic information network (GIN) model for systematically incorporating biological databases into the prioritization of SNPs after a GWAS (4). SPOT implements the GIN prioritization method using a secure, interactive web-based approach. The user uploads a list of SNPs and may optionally include P-values from statistical tests of genotype–phenotype correlation. The user may also upload a list of genes, genomic regions or specific SNPs with custom prioritization scores that determine how SPOT uses the GIN method to prioritize the SNPs. The results may be viewed directly in the web browser or downloaded in various formats. The GIN prioritization method is not designed to predict causal variants, and the prioritization results are not intended to be used to interpret the statistical significance of the GWAS results. Rather, it is intended to assist users in incorporating a broad range of biological hypotheses into the prioritization process using a transparent method of specifying their specific biological priorities and to provide results that are easily interpreted in terms of what went into the model and exactly how that information was used to prioritize the SNPs.
METHODS
Genomic Information Networks
Given a list of SNPs and P-values from a statistical test for genetic association, the goal of the GIN prioritization method (4) is to combine biological information with evidence for genetic association to prioritize SNPs for further study so that SNPs with biologically relevant properties receive higher priority. This is done by first specifying a non-negative prioritization score for each SNP. If S is the GIN prioritization score, the weighted P-value Pw is defined by Pw = P/10S (4–5). SNPs are ranked for further study by Pw where smaller values of Pw have higher priority.
A GIN prioritization score represents an order of magnitude change in P-value from a test for association. For example, a SNP with an overall score of 1 and a P-value of 0.01 has the same priority in the GIN model as a SNP with a score of 0 and P-value of 0.001. This allows the user to specify their priority for testing certain biological hypotheses by weighing that priority against evidence for genotype–phenotype correlation. Typically there are few P-values from a GWAS <10−8, so a score of ≥8 essentially guarantees a SNP will have extremely high priority after GIN prioritization. The effect of a score on rank, however, depends on the overall distribution of scores the user has specified for their set of SNPs. For example, there is no effect on rank if all SNPs receive a score of 8; the rankings are the same as by P-value alone [for more information see the discussion of normalized weights in (4)]. When deciding on a score, it is helpful to use a frame of reference. By default, we prioritize SNPs in genes one order of magnitude higher than those not in or near genes, and missense SNPs one order of magnitude higher than those in introns. In Saccone et al. (4), we conducted a sensitivity analysis to demonstrate that the GIN prioritization results are not sensitive to small changes in prioritization scores.
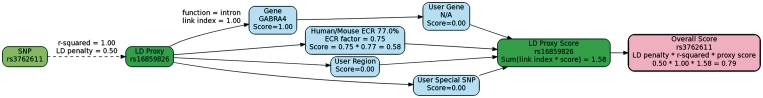
Figure 1 shows a screenshot from SPOT displaying the GIN diagram for the SNP rs3762611, which is in the example data provided on SPOT’s main page. The diagram, created dynamically by SPOT using GraphViz (http://www.graphviz.org), shows how the overall GIN prioritization score was determined for rs3762611. When determining the score, SPOT takes into account all possible LD proxies—SNPs with r2 above a certain threshold in a specific HapMap (6) sample. We used genotype data from HapMap Public Release 27 (http://hapmap.ncbi.nlm.nih.gov) and the program Haploview (7) to estimate the r2 LD coefficients in each of the 11 populations from this release. Figure 1 shows the GIN calculations for the LD proxy rs16859826, which was determined using the HapMap European American sample. The overall prioritization score is computed by traversing the network diagram from left to right and adding up scores from the different nodes as described in (4). SNPs with higher biological relevance, such as those in genes, particularly user-specified priority genes, and in conserved regions, receive higher scores.
Figure 1.
A screenshot from SPOT showing a graphical representation of the GIN for rs3762611.
The process is repeated for all LD proxies. The highest scoring LD proxy is used to determine the overall score of the original SNP, rs3762611, provided the score of the proxy is greater than the score of the original. In this case, the LD proxy rs16859826 was used to determine the overall score of rs3762611. This computation takes into account the strength of the LD proxy, so that proxies with smaller values of r2 have smaller scores and are less likely to be used. Also, a fixed penalty is applied to the score of the proxy in order to ensure LD proxies are only used when they have better scores than the original, even when r2 = 1. The default LD penalty of 0.9 is moderate so that LD proxies will be given significant attention when prioritizing SNPs. This parameter can be configured on SPOT’s main page.
Following the convention introduced in (4), the ‘Gene’ node has a score of 1 for all genes and its contribution to the overall score takes into account SNP/gene functional properties using a mechanism called the ‘link index’. In general, the link index is used to describe the strength or the manner in which a connection is made to a node. Missense mutations, for example, have a default link index of 2 and so the contribution of the ‘Gene’ node becomes 2*1 = 2. The overall score is the sum of the contributions S =∑ LiSi over the link indices Li and scores Si of the nodes. The link index of the gene node is completely determined by the user for SNP/gene transcript functional properties. These include nonsense, frameshift, missense and 5′and 3′-UTR designations. The SNP/gene transcript functional properties we used are from dbSNP build 130 (8). The 5′- and 3′-UTR specifications imply only that SNPs are located in these ends of the transcribed region; there is currently no additional information on putative promoters, transcription factor binding sites or other regulatory data.
We have incorporated information from the PolyPhen method of predicting the effect of an amino acid substitution on the properties of the protein product (9–10). PolyPhen predictions for SNPs in dbSNP build 126 were downloaded from the PolyPhen web site (http://genetics.bwh.harvard.edu/pph/data; the PolyPhen 2 tool is now available at http://genetics.bwh.harvard.edu/pph2 but at the time of writing did not yet appear to provide a comprehensive set of predictions for download). When PolyPhen predictions are enabled by the user on the main page, the SNP/gene functional property will be replaced by the PolyPhen prediction when a prediction for that SNP exists. The prediction can be ‘benign’, ‘possibly damaging’ or ‘probably damaging’, and users may specify a different prioritization score for each prediction. By default, these are 1, 2 and 3, respectively, which correspond to the default values for intron, missense and frameshift, so that the PolyPhen prediction may increase or decrease the prioritization score compared to the default value of 2 for missense SNPs.
The user may prioritize specific genes by providing gene symbols and prioritization scores as input to SPOT. This information is represented by the ‘User Gene’ node. Similarly, the user may prioritize genomic regions and single SNPs in the ‘User Region’ and ‘User Special SNP’ nodes, respectively. When a gene is specified two different ways, or when two user-specified regions overlap, SPOT will combine the prioritization scores according to the user-specified option ‘Multiple Query Method’, which may be maximum (the default), minimum, sum or average. When a SNP is in an evolutionary conserved region (ECR) with fractional conservation P (0 ≤ P ≤ 1), the corresponding ECR node contributes F*P to the overall score where F is a factor currently set to 0.75. We used ECRBase for data on human/mouse ECRs (11).
The final results of the GIN prioritization process may be viewed in an interactive table within the web browser (see the SPOT User’s Guide, https://spot.cgsmd.isi.edu/doc/user_guide.pdf, for screenshots and additional details). The table shows the original P-values and their ranks in the column ‘Rank: p-value’, and the SPOT rankings from the GIN prioritization method in ‘Rank: SPOT’ which are determined by the ‘GIN weighted p-value’ column. The ‘Rank: SPOT’ column is the most important item in the table as it reflects the priority of the SNP from the GIN prioritization method. A graphical column selection tool shown can be used to configure the output tables. The user may select which columns are displayed and their order. This allows the user to view additional information such as detailed gene/mapping properties, HapMap allele frequencies and commercial SNP microarray LD tagging properties. Columns labeled with an asterisk refer to the LD proxy when one is being used, and information about the original or ‘source’ SNP may be viewed using the column selection tool.
Using SPOT
The main page consists of a web form for the primary user input. Users must first either upload a list of numeric SNP identification numbers as a file or enter the list directly into a web form. The user may optionally provide P-values from statistical tests for genotype–phenotype correlation. In the section labeled ‘Prioritization of specific genes and other genomic regions’, the user can specify a list of genes and prioritization scores to be used in the ‘User Gene’ node of the GIN model.
A number of methods can be used to specify genes and genomic regions. For example, the query ‘ENTREZ_GENE_QUERY = Dopamine and Receptor,1.0’retrieves all genes from the Entrez Gene database (http://www.ncbi.nlm.nih.gov/sites/entrez?db=gene) that match the terms ‘dopamine’ and ‘receptor’ and assigns them all a prioritization score of 1.0. SNPs from an entire region can be prioritized with a query of the form ‘REGION = Chr15:76600000.76700000, 1.25′, which would add a score of 1.25 to all SNPs in this 100 Kb region on chromosome 15 via the ‘User Region’ node of the GIN model. More information on the different kinds of queries is provided in the ‘User’s Guide’ (https://spot.cgsmd.isi.edu/doc/user_guide.pdf).
Programmatic architecture
MySQL (http://www.mysql.com) is used via the Perl DBI module (http://dbi.perl.org) to process and store user input and implement the GIN prioritization method. During this execution, biological information is integrated from a local MySQL relational database. All results are stored as tables in a MySQL relational database which is then accessed by the web interface.
SPOT’s web interface makes use of different technologies and programming languages. From the user perspective, it only requires any modern web browser such as Firefox, Internet Explorer, Chrome, Safari or Opera. Mainly it is a web application developed with XHTML, JavaScript and CSS at the client-side (AJAX) and PHP/Perl at the server-side. It respects the model view controller paradigm and isolates the user interface from the logic and the data of the application. The JavaScript codes reuse and take advantage of several JavaScript libraries such as jQuery (http://jquery.com), Prototype (http://prototypejs.org), jqGrid (http://www.trirand.com/blog) and their plug-ins. Then the web user interface allows the user to navigate through the MySQL database created by the Perl script. We have created a Wiki for SPOT (http://confluence.pegasus.isi.edu/display/CGSMD/Howto+install+SPOT) with detailed technical information and resources for downloading and installing a local copy of SPOT, including the underlying MySQL relational database of biological information.
Execution time
In the following runs of SPOT, the default LD settings were used so that LD proxies from the HapMap CEU (European–American) sample are used with a threshold of r2 ≥ 0.8. The default setting of a maximum of 1000 SNPs limits the number of SNPs sent to the output tables, but does not limit SNPs used as input, and the P-value threshold is set to 0.05 so that only SNPs with P ≤ 0.05 are used in the GIN model. The example data provided on the main page (10 SNPs and 4 queries retrieving 10 genes) took 3 s to run, and a more typical run of 1 million SNPs with randomly selected, and therefore uniformly distributed, P-values and 1000 prioritized genes took 57 s (in this case, about 50 000 SNPs were used for prioritization given the P ≤ 0.05 filter). A more ambitious run of 1 million SNPs and 10 000 prioritized genes took 3 min and 13 s.
Security
The results of a GWAS are sensitive information and SPOT takes several steps to ensure this information is protected during a session and is destroyed when the session is complete. SPOT uses a DigiCert encryption certificate (http://www.digicert.com) so that all communication between the user and the server are secure. Any information the user uploads is destroyed after 3 h or immediately at the user’s request. Since the results depend only on the relative order of the SNPs, the user may scale the P-values by a fixed factor prior to uploading them so that the true values are never transmitted. Nevertheless, if the user does not wish to upload P-values they may select the option ‘SNPs only, no p-values’ on the main page. In this case, SPOT provides an Excel file with the prioritization scores into which the P-values can be pasted. SPOT includes a calculated column in the Excel file that computes the weighted P-values (Pw = P/10S where S is the overall prioritization score) which are used to determine the prioritized rankings from the GIN method. This process takes place on the user’s computer so that the results are obtained without ever transferring P-values to the SPOT server.
FUTURE DEVELOPMENT
The GIN prioritization method is designed to be flexible and allow a variety of biological databases to be incorporated while remaining viable and interpretable. Future iterations of SPOT will include additional biological information such as expression quantitative trait loci (12), transcription factor binding sites, micro RNA target sites and other GWAS results [see (13) and (14) for some proposed methods on implementing these kinds of data]. A useful feature would be to add predefined gene sets to SPOT’s query tool. For example, the NeuroSNP database (3) (http://nidagenetics.org/neurosnp/index.htm) consists of genes related to addiction-related diseases. Future iterations will allow the user to specify such a database, either with predefined prioritization scores or a user-defined score for the entire set. This feature would be added for convenience only as users can currently accomplish this by directly entering these gene lists into SPOT. Similar to the idea behind disease-related gene databases is the Human Variome Project (15) that aims to develop novel methods of cataloging human genetic variation and its relation to disease, and like gene databases the results of this project could be incorporated directly into SPOT. As next generation sequencing becomes the standard in following up on GWAS, the discovery and analysis of numerous and possibly very important rare variants (16) may require biological prioritization to design follow-up studies, and we will be studying ways of modifying SPOT to deal with this challenging problem.
DISCUSSION
SPOT is designed to provide investigators with a tool for systematically incorporating their specific biological hypotheses into the post-GWAS prioritization process. The testing of biological hypotheses is a reasonable study design, even if there is no clear evidence of a predictive mechanism. While a GWAS is often touted as being ‘hypothesis free’, this is not exactly the case. Typically a GWAS tests only a particular subset of variants, and designs differ from array to array with some arrays taking into account biological information (3,17). Prior to the availability of GWAS technology, researchers tested candidate genes—a logical and reasonable experiment given the available resources whose success or failure moves the field closer to the goal of testing all known variants with acceptable statistical power. Other examples of biological study designs are GWAS using only non-synonymous SNPs (18,19) and exome sequencing (20). One of the advantages of studying variants with clear biological, although possibly not phenotypically causal, consequences, such as missense SNPs as compared to SNPs not in or near genes, is the potential for conducting functional studies such as knockouts in animal models (21). SPOT mimics these study designs by allowing researchers to specify the particular biological hypotheses they wish to test so that SNPs related to these hypotheses receive additional priority when resources are limited. As has been submitted elsewhere in the literature (1,5,22), this strategy is reasonable with the stipulation that biological priorities should be established a priori in order to avoid post hoc arguments for biological plausibility, because given a gene it is not difficult to use Internet databases to mine connections between that gene and a phenotype. As we described in (4), the GIN prioritization method is well suited for establishing a specific, quantitative a priori plan for implementing biological hypotheses into the post-GWAS prioritization process, and this method is now implemented in SPOT.
SPOT is a useful tool for GWAS investigators because it allows them to test these reasonable biological hypotheses by defining specific biological priorities as a way of pursuing the ‘high hanging fruit’ in a GWAS. A GWAS often has limited statistical power and must rely on replication genotyping to establish the statistical significance of any remaining non-significant associations that appear promising. Even when GWAS experiments successfully replicate SNP associations, the totality of these variants often explain only a fraction of the genetic variance (23). We then look to the ‘high hanging fruit’ for answers—variants with smaller effect sizes that require greater power to be confirmed as a statistically significant association (23). These may require substantial further investigation such as large meta-analyses conducted by consortia with sample sizes often in the hundreds of thousands (24–26). It has also been argued that even with missing variance, one of the benefits of GWAS is, in addition to predicting individual risk, their ability to expose biological pathways that underlie human disease (2,27). Given the extremely high resource-consuming nature of these follow-up studies, a clear and precise plan for the prioritization of variants is critical. Clearly those variants with the strongest statistical evidence of association will be pursued first, but as that evidence dwindles, some signals with no evidence of biological relevance may be traded for those meeting the biological priorities of the study when the difference in evidence for genotype–phenotype correlation is modest. As shown by Saccone et al. (4), this is what occurs when the GIN prioritization method is implemented after a GWAS: the difference between the set of SNPs selected by the GIN prioritization and straight P-value methods is roughly only one order of magnitude in association with P-value. The difference applies mainly to SNPs with moderate evidence for association—the potential ‘high hanging fruit’.
An example of biological prioritization that could be interpreted as a ‘success’ is the following discovery of a novel genetic association with nicotine dependence. In Saccone et al. (28) the investigators conducted a large candidate gene study of nicotine dependence. Although there were no statistically significant results after correcting for multiple testing, the fifth smallest P-value was the missense SNP rs16969968 in the nicotinic receptor gene CHRNA5 and was highlighted as the most promising signal. A GWAS of nicotine dependence (29) was conducted in conjunction with the candidate gene study, and although the missense SNP ranked 199 in the combined GWAS/candidate gene results, it was the top priority for further study in the overall project. It has since been replicated in numerous studies of nicotine dependence, heavy smoking, lung cancer and COPD (30–36), including three very large meta-analytic studies (24–26). Furthermore, there have been very few replicated associations other than the original SNP and some SNPs in nearby genes. Clearly this example alone is not evidence of the general predictive power of biological information, in this case a missense SNP in a gene whose protein product binds to the drug of interest, to predict true genetic association. Nevertheless, the results from a study by Saccone et al. (3) show that commercial SNP microarrays may miss a significant amount of coverage in some genes. The fact that an overall GWAS could be negative due to the omission of a single SNP that could be discovered by another study targeting a small number of highly biologically relevant SNPs [rs16969968 was in the top 10 over all of dbSNP when we used the GIN method to prioritize for nicotine dependence (4)] is something for GWAS researchers to consider, both for the post-GWAS selection of SNPs for further study and for the pre-GWAS supplementation of commercial arrays (3).
SPOT is not intended to be used to predict true causal variants or to statistically interpret the results of GWAS. Furthermore, the problem of establishing such predictive properties for SPOT, such as through assessments of false-positive rates and receiver operating characteristics, is ill posed due to the fact that one of the core features of SPOT is that it allows the investigator to prioritize specific genes and genomic regions. Since these parameters depend on the particular biological priorities of the investigator, the general predictive properties of SPOT cannot be established. Another issue is the extreme diversity of phenotype and disease. For example, few genetic associations for psychiatric disease have been validated by replication in independent samples (37). Therefore, in order to conclude that any evidence of correlation between the biological information used by SPOT and existing confirmed genotype–phenotype associations would transfer to the prediction of true genetic associations for psychiatric disease, one would have to make the unlikely assumption that the underlying genetic structure of psychiatric disease shares substantial common elements with other types of human disease in general.
A third problem in assessing the predictive properties of SPOT is the extraordinary challenge of assembling a sufficiently sized collection of validated ‘true’ causal variants for common complex disease on which an assessment of prediction must be based. Even after ‘true’ genotype–phenotype correlations have been validated using the rigorous standards initiated by the onset of GWAS, including statistical significance after correcting for genome-wide multiple testing (1), proper adjustment for population stratification verified by an acceptable genomic inflation factor (18) and replication in independent studies (1), a critical issue that remains is distinguishing the actual causal variants from a potentially large number of LD proxies (1,38). Association statistics are virtually indistinguishable for SNPs in strong LD so that the actual causal variants must be identified by other means such as functional studies. The problem is that the result of a prediction algorithm could be ‘positive’ for an associated non-causal SNP and ‘negative’ for a causal LD proxy. This could artificially inflate the estimate of prediction. The confirmation of pathogenesis may be more straightforward for highly penetrant mutations causing rare Mendelian disorders. Tools that predict the effect of amino acid substitutions, such as PolyPhen (10) and SIFT (9,39), have been shown to have predictive power for Mendelian disorders. PolyPhen predictions have been incorporated into SPOT and their influence on priority can be configured by the user. GWAS, however, are more directly aimed at common complex disease (40,41), and it is an enormous challenge to assemble a collection of causal variants for common complex disease that meets these rigorous criteria for validation, in particular the disambiguation of LD proxies, that is of sufficient size to assess predictive power. While projects such as GEN2PHEN (http://www.gen2phen.org) and the Human Variome Project (15) aim to solve this problem [see (42) for a general review], it would appear that currently the resource closest to meeting these validation criteria is the database maintained by the National Human Genome Research Institute containing published GWAS associations with P < 10−5 from the statistical test for association (http://www.genome.gov/gwastudies/) (38), although this information does not resolve the LD disambiguation issue.
Since the SPOT prioritization results are not intended to predict causal variants, we recommend that the statistical significance of GWAS results be evaluated based on genotype–phenotype correlation data alone and be corrected for genome-wide multiple testing in accordance with current standards (1). In particular, this practice will help to guard against bias from reports of positive association in the literature, as well a bias from the biological priorities of the investigators which may lead to a misinterpretation of the results of the GWAS.
At the time of writing, there are very few publicly accessible web-based tools that perform the same function as SPOT, namely taking GWAS results as input and as output providing a table with rankings that take into account user-defined measures of biological relevance. A number of web tools dealing with SNP biological properties are shown in Table 1. GenePipe (43) is the closet to SPOT in purpose and functionality. It takes association results as input, as well as user-defined weights for various forms of genomic annotation, and provides an annotated table as output. Currently, GenePipe incorporates more databases while SPOT offers greater transparency in conveying the prioritization method by providing side by side GIN versus P-value rankings, graphical representations of the GIN calculations and tables showing the details of the prioritization process step by step, all presented interactively in the web browser.
Table 1.
Some web tools dealing with SNP biological properties and their characteristics related to the prioritization of GWAS results
| Web tool | Exclusively for non-synonymous SNPs | Accepts multiple SNPs | Accepts P-values | Performs customizable GWAS prioritization | Incorporates LD proxies | Some external data sources used (Table 2) |
|---|---|---|---|---|---|---|
| F-SNP (44) | No | No | No | No | No | 1,4,5,6,9–20 |
| FastSNP (45) | No | Yes | No | No | No | 2,4,5,7,8,13,14,19 |
| GenePipe (43) | No | Yes | Yes | Yes | Yes | 2,8,13,14,17,20 |
| Panthera (46) | Yes | Noa | No | No | No | 4 |
| PolyPhen2 (9–10) | Yes | Yes | No | No | No | 2 |
| SIFT (39) | Yes | Yes | No | No | No | 2 |
| SNPs3Db (47) | Yes | Noc | No | No | No | 2 |
| SPOT (4) | No | Yes | Yes | Yes | Yes | 2,3,8,13 |
The values in the column ‘External data sources used’ refer to the ‘Number’ column in Table 2; this list of external data sources used may not be complete.
aThis is the cSNP web tool on the Panther site. The site states that a tool can be downloaded for analyzing multiple SNPs.
bWe studied the tool labeled ‘Impact on Protein Structure and Function’ on the SNPs3D main page.
cSNPs3D does offer an ‘annotate SNPs’ feature that apparently requires registration and we did not explore this.
In addition to GenePipe, there are a number of other web tools such as F-SNP (44), FastSNP (45), Panther (46), PolyPhen2 (9–10), SIFT (39) and SNPs3D (47) which assess the biological relevance of a SNP independently of genotype–phenotype correlation results (Table 1). Half of the eight tools shown in Table 1 deal exclusively with non-synonymous SNPs. Of the remaining four, only SPOT and FastSNP attempt to combine evidence from multiple sources of information and return a single measure of biological relevance suitable for systematically prioritizing GWAS results. FastSNP, while providing a great deal of information in very informative diagrammatic format, does not account for LD proxies. The evidence that the tools in Table 1 can be used to predict causal variants that influence general common complex disease, subject to the aforementioned validation criteria for causal variants, is limited. PolyPhen (10) and SIFT (39), which deal exclusively with non-synonymous SNPs, have found evidence of prediction using information on disease-related variants from the UniProt database (60) for Mendelian disorders flagged by terms such as ‘lethal’ and ‘complete loss of function’ (PolyPhen is incorporated into SPOT). FastSNP and GenePipe, which can be used for arbitrary SNPs, each tested the predictive properties based on a single disease study. A feature of some of these other tools that may be significantly appealing to investigators when compared to SPOT is the fact that they offer substantially more biological information, as shown in Tables 1 and 2. However, with the exception of GenePipe, due to limited input features these tools appear to be more geared towards assessing the biological plausibility of a small number of SNPs and perhaps their LD proxies (although these would have to be obtained from a different source, such as the LD web tool SNAP (61), and submitted manually). This could be applied to a small number of ‘top hits’ from a GWAS, as opposed to incorporating biological information into a full-scale GWAS for the purpose of prioritizing a large number of replication experiments, which is the purpose of SPOT. We are currently exploring ways of integrating additional biological information, such as from the sources in Tables 1 and 2, in order to provide users with a more comprehensive palette for establishing biological hypotheses.
Table 2.
Some data sources used by the web tools in Table 1 based on the latest documentation from their web sites and the corresponding bibliographic citations
| Number | Name | Description |
|---|---|---|
| 1 | Consitea (48) | Conserved transcription factor binding sites |
| 2 | dbSNP (8) | General SNP/gene transcript properties |
| 3 | ECRBasea (11) | Evolutionary conserved regions |
| 4 | Ensembl (49) | Extensive genomic database including SNPs and gene transcripts |
| 5 | ESEfindera (50) | Exonic splice sites |
| 6 | ESRSearch (51) | Exonic-splicing regulatory (ESR) sequences |
| 7 | FAS-ESSa (52) | Predicts exonic splicing silencer for each SNP allele |
| 8 | HapMap (6) | Dense genotyping on multiple populations, useful for LD estimates |
| 9 | KinasePhosa (53) | Phosphorylation sites |
| 10 | LS-SNP (54) | SNP annotation tool |
| 11 | OGPETa,b | Prediction of O-glycosylation sites in proteins |
| 12 | PESXa (55) | Exon splicing enhancers/silencers |
| 13 | PolyPhen (9–10) | Prediction of amino acid substitution effects |
| 14 | RescueESEa (56) | Exonic splice sites |
| 15 | SIFT (39) | Prediction of amino acid substitution effects |
| 16 | SNPeffect (57) | SNP annotation with human disease |
| 17 | SNPs3D (47) | Impact of nsSNPs on protein function |
| 18 | Sulfinatora (58) | Tyrosine sulfination sites |
| 19 | TFSearcha,b | Transcription factor binding sites |
| 20 | UCSC (59) | Extensive genomic database including SNPs and gene transcripts |
aAt the time of writing, this site did not accept dbSNP reference SNP, or ‘rs’, identification numbers as input.
bAt the time of writing, we were unable to locate a publication related to this resource. See the ‘Acknowledgements’ Section for more information.
In Saccone et al. (4), we conducted a sensitivity analysis of how changes in the user’s prioritization scores effect the rankings, and we performed simulations to assess the difference between selecting SNPs after a GWAS using just P-value rankings and using the GIN method. Neither of these analyses was designed to use a ‘training set’ to establish predictive properties. We found the rankings are not very sensitive to changes in the scores and that in general with default scoring parameters there is about a one order of magnitude difference between SNPs selected by P-value and those selected by the GIN method.
The SNP/gene transcript properties used by our algorithm are currently limited to a sophisticated prediction provided by the PolyPhen (9–10) algorithm on the impact of an amino acid substitution, and those that can be observed directly from DNA and RNA sequence such as coding regions, untranslated regions, missense and nonsense amino acid substitutions and frameshifts. Given the amount of experimental human genomic data available, this is a relatively limited amount of information. However, when integrating biological information into a GWAS, even with this relatively limited set, there are several aspects to consider. First, a SNP may be associated with many genes, whether it be in one gene and near another, or in the intersection of multiple genes or perhaps in a gene with several known transcripts due to alternative splicing and having different functional consequences of the SNP on each transcript. SPOT considers all known SNP/gene transcript associations, and selects the one with the highest priority to ensure no biologically promising association signal is missed. Furthermore, when genes overlap, SPOT will take into account specific genes prioritized by the user. Finally, the task of checking for LD proxies while taking into account all of the previous elements is not only a complex algorithm to implement, but requires the processing of an enormous amount of information (our LD database alone contains data on 343 million pairs of SNPs) that must be implemented on a genome-wide scale when applied to GWAS data. While we are planning to incorporate additional biological information into future implementations of SPOT, researchers should find SPOT useful even with this relatively limited amount of information.
The problem of interpreting the results of a GWAS and planning follow-up experiments is formidable. Integrating information from biological databases can aid the decision making process in order to maximize resources. SPOT is an easily implemented and flexible tool that will aid researchers in applying a biological prioritization strategy when selecting SNPs for further study after a GWAS and is designed to remain an interpretable and viable solution as additional sources of biological information are integrated.
FUNDING
National Institutes of Health (DA024722 to S.F.S., MH068457 to J.A.T., AA008401 to J.P.R., NIDA Contract HHSN271200900012C to J.A.T.); American Cancer Society (IRG5801050 to S.F.S.). Funding for open access charge: National Institutes of Health Grant DA024722.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We are very grateful to the following individuals for testing SPOT: William Howells, Chun-Nan Hsu, Peng Lin, Richard C. McEachin, Nanette Rochberg, Sharon Ryan, Nancy L. Saccone and Andrew Schrage. We would also like to thank Gary Stormo for valuable advice concerning certain aspects of the design. OGPET was developed by Rafael Torres Jr., Yash Dayal, Ming-Ying Leung, and Igor C. Almeida, Border Biomedical Research Center (BBRC), Departments of Biological and Mathematical Sciences, University of Texas at El Paso. Finally, we are very appreciative of the time and effort invested by the reviewers, whose comments and suggestions resulted in numerous improvements to the manuscript and enhancements to the web tool itself.
REFERENCES
- 1.Chanock SJ, Manolio T, Boehnke M, Boerwinkle E, Hunter DJ, Thomas G, Hirschhorn JN, Abecasis G, Altshuler D, Bailey-Wilson JE, et al. Replicating genotype-phenotype associations. Nature. 2007;447:655–660. doi: 10.1038/447655a. [DOI] [PubMed] [Google Scholar]
- 2.Cantor RM, Lange K, Sinsheimer JS. Prioritizing GWAS results: a review of statistical methods and recommendations for their application. Am. J. Hum. Genet. 2010;86:6–22. doi: 10.1016/j.ajhg.2009.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Saccone SF, Bierut LJ, Chesler EJ, Kalivas PW, Lerman C, Saccone NL, Uhl GR, Li CY, Philip VM, Edenberg HJ, et al. Supplementing high-density SNP microarrays for additional coverage of disease-related genes: addiction as a paradigm. PLoS ONE. 2009;4:e5225. doi: 10.1371/journal.pone.0005225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Saccone SF, Saccone NL, Swan GE, Madden PA, Goate AM, Rice JP, Bierut LJ. Systematic biological prioritization after a genome-wide association study: an application to nicotine dependence. Bioinformatics. 2008;24:1805–1811. doi: 10.1093/bioinformatics/btn315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Roeder K, Bacanu SA, Wasserman L, Devlin B. Using linkage genome scans to improve power of association in genome scans. Am. J. Hum. Genet. 2006;78:243–252. doi: 10.1086/500026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Frazer KA, Ballinger DG, Cox DR, Hinds DA, Stuve LL, Gibbs RA, Belmont JW, Boudreau A, Hardenbol P, Leal SM, et al. A second generation human haplotype map of over 3.1 million SNPs. Nature. 2007;449:851–861. doi: 10.1038/nature06258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 8.Sayers EW, Barrett T, Benson DA, Bolton E, Bryant SH, Canese K, Chetvernin V, Church DM, Dicuccio M, Federhen S, et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2010;38:D5–D16. doi: 10.1093/nar/gkp967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ramensky V, Bork P, Sunyaev S. Human non-synonymous SNPs: server and survey. Nucleic Acids Res. 2002;30:3894–3900. doi: 10.1093/nar/gkf493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, Kondrashov AS, Sunyaev SR. A method and server for predicting damaging missense mutations. Nat. Methods. 2010;7:248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Loots G, Ovcharenko I. ECRbase: database of evolutionary conserved regions, promoters, and transcription factor binding sites in vertebrate genomes. Bioinformatics. 2007;23:122–124. doi: 10.1093/bioinformatics/btl546. [DOI] [PubMed] [Google Scholar]
- 12.Nicolae DL, Gamazon E, Zhang W, Duan S, Dolan ME, Cox NJ. Trait-associated SNPs are more likely to be eQTLs: annotation to enhance discovery from GWAS. PLoS Genet. 2010;6:e1000888. doi: 10.1371/journal.pgen.1000888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Barnes MR, Plumpton M. Predictive functional analysis of polymorphisms: an overview. In: In Barnes MR, editor. Bioinformatics for Geneticists. 2nd edn. West Sussex, England: John Wiley & Sons, Ltd; 2007. pp. 249–280. [Google Scholar]
- 14.Barnes MR, Derwent PS. Needle in a haystack? Dealing with 500,000 SNP genome scans. In: Barnes MR, editor. Bioinformatics for Geneticists. 2nd edn. West Sussex, England: John Wiley & Sons, Ltd; 2007. pp. 447–493. [Google Scholar]
- 15.Howard HJ, Horaitis O, Cotton RG, Vihinen M, Dalgleish R, Robinson P, Brookes AJ, Axton M, Hoffmann R, Tuffery-Giraud S. The Human Variome Project (HVP) 2009 Forum “Towards Establishing Standards”. Hum. Mutat. 2010;31:366–367. doi: 10.1002/humu.21175. [DOI] [PubMed] [Google Scholar]
- 16.Dickson SP, Wang K, Krantz I, Hakonarson H, Goldstein DB. Rare variants create synthetic genome-wide associations. PLoS Biol. 2010;8:e1000294. doi: 10.1371/journal.pbio.1000294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kitsios GD, Zintzaras E. Genome-wide association studies: hypothesis-“free” or “engaged”? Trans. Res. 2009;154:161–164. doi: 10.1016/j.trsl.2009.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Clayton DG, Walker NM, Smyth DJ, Pask R, Cooper JD, Maier LM, Smink LJ, Lam AC, Ovington NR, Stevens HE, et al. Population structure, differential bias and genomic control in a large-scale, case-control association study. Nat. Genet. 2005;37:1243–1246. doi: 10.1038/ng1653. [DOI] [PubMed] [Google Scholar]
- 19.Hampe J, Franke A, Rosenstiel P, Till A, Teuber M, Huse K, Albrecht M, Mayr G, De La Vega FM, Briggs J, et al. A genome-wide association scan of nonsynonymous SNPs identifies a susceptibility variant for Crohn disease in ATG16L1. Nat. Genet. 2007;39:207–211. doi: 10.1038/ng1954. [DOI] [PubMed] [Google Scholar]
- 20.Ng SB, Buckingham KJ, Lee C, Bigham AW, Tabor HK, Dent KM, Huff CD, Shannon PT, Jabs EW, Nickerson DA, et al. Exome sequencing identifies the cause of a mendelian disorder. Nat. Genet. 2010;42:30–35. doi: 10.1038/ng.499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Portugal GS, Gould TJ. Genetic variability in nicotinic acetylcholine receptors and nicotine addiction: converging evidence from human and animal research. Behav. Brain Res. 2008;193:1–16. doi: 10.1016/j.bbr.2008.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wacholder S, Chanock S, Garcia-Closas M, El Ghormli L, Rothman N. Assessing the probability that a positive report is false: an approach for molecular epidemiology studies. J. Natl Cancer Inst. 2004;96:434–442. doi: 10.1093/jnci/djh075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Goldstein DB. Common genetic variation and human traits. N. Engl. J. Med. 2009;360:1696–1698. doi: 10.1056/NEJMp0806284. [DOI] [PubMed] [Google Scholar]
- 24.Furberg H, Kim Y, Dackor J, Boerwinkle E, Franceschini N, Ardissino D, Bernardinelli L, Mannucci PM, Mauri F, Merlini PA, et al. Genome-wide meta-analyses identify multiple loci associated with smoking behavior. Nat. Genet. 2010;42:441–447. doi: 10.1038/ng.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thorgeirsson TE, Gudbjartsson DF, Surakka I, Vink JM, Amin N, Geller F, Sulem P, Rafnar T, Esko T, Walter S, et al. Sequence variants at CHRNB3-CHRNA6 and CYP2A6 affect smoking behavior. Nat. genet. 2010;42:448–453. doi: 10.1038/ng.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu JZ, Tozzi F, Waterworth DM, Pillai SG, Muglia P, Middleton L, Berrettini W, Knouff CW, Yuan X, Waeber G, et al. Meta-analysis and imputation refines the association of 15q25 with smoking quantity. Nat. Genet. 2010;42:436–440. doi: 10.1038/ng.572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hirschhorn JN. Genomewide association studies–illuminating biologic pathways. N. Engl. J. Med. 2009;360:1699–1701. doi: 10.1056/NEJMp0808934. [DOI] [PubMed] [Google Scholar]
- 28.Saccone SF, Hinrichs AL, Saccone NL, Chase GA, Konvicka K, Madden PA, Breslau N, Johnson EO, Hatsukami D, Pomerleau O, et al. Cholinergic nicotinic receptor genes implicated in a nicotine dependence association study targeting 348 candidate genes with 3713 SNPs. Hum. Mol. Genet. 2007;16:36–49. doi: 10.1093/hmg/ddl438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bierut LJ, Madden PA, Breslau N, Johnson EO, Hatsukami D, Pomerleau OF, Swan GE, Rutter J, Bertelsen S, Fox L, et al. Novel genes identified in a high-density genome wide association study for nicotine dependence. Hum. Mol. Genet. 2007;16:24–35. doi: 10.1093/hmg/ddl441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Saccone NL, Wang JC, Breslau N, Johnson EO, Hatsukami D, Saccone SF, Grucza RA, Sun L, Duan W, Budde J, et al. The CHRNA5-CHRNA3-CHRNB4 nicotinic receptor subunit gene cluster affects risk for nicotine dependence in African-Americans and in European-Americans. Cancer Res. 2009;69:6848–6856. doi: 10.1158/0008-5472.CAN-09-0786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pillai SG, Ge D, Zhu G, Kong X, Shianna KV, Need AC, Feng S, Hersh CP, Bakke P, Gulsvik A, et al. A genome-wide association study in chronic obstructive pulmonary disease (COPD): identification of two major susceptibility loci. PLoS Genet. 2009;5:e1000421. doi: 10.1371/journal.pgen.1000421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Caporaso N, Gu F, Chatterjee N, Sheng-Chih J, Yu K, Yeager M, Chen C, Jacobs K, Wheeler W, Landi MT, et al. Genome-wide and candidate gene association study of cigarette smoking behaviors. PLoS ONE. 2009;4:e4653. doi: 10.1371/journal.pone.0004653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stevens VL, Bierut LJ, Talbot JT, Wang JC, Sun J, Hinrichs AL, Thun MJ, Goate A, Calle EE. Nicotinic receptor gene variants influence susceptibility to heavy smoking. Cancer Epidemiol. Biomarkers Prev. 2008;17:3517–3525. doi: 10.1158/1055-9965.EPI-08-0585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hung RJ, McKay JD, Gaborieau V, Boffetta P, Hashibe M, Zaridze D, Mukeria A, Szeszenia-Dabrowska N, Lissowska J, Rudnai P, et al. A susceptibility locus for lung cancer maps to nicotinic acetylcholine receptor subunit genes on 15q25. Nature. 2008;452:633–637. doi: 10.1038/nature06885. [DOI] [PubMed] [Google Scholar]
- 35.Chanock SJ, Hunter DJ. Genomics: when the smoke clears. Nature. 2008;452:537–538. doi: 10.1038/452537a. [DOI] [PubMed] [Google Scholar]
- 36.Berrettini W, Yuan X, Tozzi F, Song K, Francks C, Chilcoat H, Waterworth D, Muglia P, Mooser V. alpha-5/alpha-3 nicotinic receptor subunit alleles increase risk for heavy smoking. Mol. Psychiatry. 2008;13:368–373. doi: 10.1038/sj.mp.4002154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bosker FJ, Hartman CA, Nolte IM, Prins BP, Terpstra P, Posthuma D, van Veen T, Willemsen G, Derijk RH, de Geus EJ, et al. Poor replication of candidate genes for major depressive disorder using genome-wide association data. Mol. Psychiatry. 2010 doi: 10.1038/mp.2010.38. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 38.Hindorff LA, Sethupathy P, Junkins HA, Ramos EM, Mehta JP, Collins FS, Manolio TA. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc. Natl Acad. Sci. USA. 2009;106:9362–9367. doi: 10.1073/pnas.0903103106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kumar P, Henikoff S, Ng PC. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat. Protoc. 2009;4:1073–1081. doi: 10.1038/nprot.2009.86. [DOI] [PubMed] [Google Scholar]
- 40.Manolio TA, Collins FS, Cox NJ, Goldstein DB, Hindorff LA, Hunter DJ, McCarthy MI, Ramos EM, Cardon LR, Chakravarti A, et al. Finding the missing heritability of complex diseases. Nature. 2009;461:747–753. doi: 10.1038/nature08494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hardy J, Singleton A. Genomewide association studies and human disease. N. Engl J. Med. 2009;360:1759–1768. doi: 10.1056/NEJMra0808700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Thorisson GA, Muilu J, Brookes AJ. Genotype-phenotype databases: challenges and solutions for the post-genomic era. Nat. Rev. Genet. 2009;10:9–18. doi: 10.1038/nrg2483. [DOI] [PubMed] [Google Scholar]
- 43.Xu Z, Taylor JA. SNPinfo: integrating GWAS and candidate gene information into functional SNP selection for genetic association studies. Nucleic Acids Res. 2009;37:W600–W605. doi: 10.1093/nar/gkp290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lee PH, Shatkay H. F-SNP: computationally predicted functional SNPs for disease association studies. Nucleic Acids Res. 2007;36:D820–D824. doi: 10.1093/nar/gkm904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yuan HY, Chiou JJ, Tseng WH, Liu CH, Liu CK, Lin YJ, Wang HH, Yao A, Chen YT, Hsu CN. FASTSNP: an always up-to-date and extendable service for SNP function analysis and prioritization. Nucleic Acids Res. 2006;34:W635–W641. doi: 10.1093/nar/gkl236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mi H, Dong Q, Muruganujan A, Gaudet P, Lewis S, Thomas PD. PANTHER version 7: improved phylogenetic trees, orthologs and collaboration with the Gene Ontology Consortium. Nucleic Acids Res. 2010;38:D204–D210. doi: 10.1093/nar/gkp1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yue P, Melamud E, Moult J. SNPs3D: candidate gene and SNP selection for association studies. BMC Bioinformatics. 2006;7:166. doi: 10.1186/1471-2105-7-166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sandelin A, Wasserman WW, Lenhard B. ConSite: web-based prediction of regulatory elements using cross-species comparison. Nucleic Acids Res. 2004;32:W249–W252. doi: 10.1093/nar/gkh372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hubbard TJ, Aken BL, Ayling S, Ballester B, Beal K, Bragin E, Brent S, Chen Y, Clapham P, Clarke L, et al. Ensembl 2009. Nucleic Acids Res. 2008;37:D690–D697. doi: 10.1093/nar/gkn828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cartegni L, Wang J, Zhu Z, Zhang MQ, Krainer AR. ESEfinder: a web resource to identify exonic splicing enhancers. Nucleic Acids Res. 2003;31:3568–3571. doi: 10.1093/nar/gkg616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Goren A, Ram O, Amit M, Keren H, Lev-Maor G, Vig I, Pupko T, Ast G. Comparative analysis identifies exonic splicing regulatory sequences–the complex definition of enhancers and silencers. Mol. cell. 2006;22:769–781. doi: 10.1016/j.molcel.2006.05.008. [DOI] [PubMed] [Google Scholar]
- 52.Wang Z, Rolish ME, Yeo G, Tung V, Mawson M, Burge CB. Systematic identification and analysis of exonic splicing silencers. Cell. 2004;119:831–845. doi: 10.1016/j.cell.2004.11.010. [DOI] [PubMed] [Google Scholar]
- 53.Wong YH, Lee TY, Liang HK, Huang CM, Wang TY, Yang YH, Chu CH, Huang HD, Ko MT, Hwang JK. KinasePhos 2.0: a web server for identifying protein kinase-specific phosphorylation sites based on sequences and coupling patterns. Nucleic Acids Res. 2007;35:W588–W594. doi: 10.1093/nar/gkm322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ryan M, Diekhans M, Lien S, Liu Y, Karchin R. LS-SNP/PDB: annotated non-synonymous SNPs mapped to Protein Data Bank structures. Bioinformatics. 2009;25:1431–1432. doi: 10.1093/bioinformatics/btp242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhang XH, Chasin LA. Computational definition of sequence motifs governing constitutive exon splicing. Genes Dev. 2004;18:1241–1250. doi: 10.1101/gad.1195304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fairbrother WG, Yeh RF, Sharp PA, Burge B. Predictive identification of exonic splicing enhancers in human genes. Science. 2002;297:1007–1013. doi: 10.1126/science.1073774. [DOI] [PubMed] [Google Scholar]
- 57.Reumers J, Maurer-Stroh S, Schymkowitz J, Rousseau F. SNPeffect v2.0: a new step in investigating the molecular phenotypic effects of human non-synonymous SNPs. Bioinformatics. 2006;22:2183–2185. doi: 10.1093/bioinformatics/btl348. [DOI] [PubMed] [Google Scholar]
- 58.Monigatti F, Gasteiger E, Bairoch A, Jung E. The Sulfinator: predicting tyrosine sulfation sites in protein sequences. Bioinformatics. 2002;18:769–770. doi: 10.1093/bioinformatics/18.5.769. [DOI] [PubMed] [Google Scholar]
- 59.Rhead B, Karolchik D, Kuhn RM, Hinrichs AS, Zweig AS, Fujita PA, Diekhans M, Smith KE, Rosenbloom KR, Raney BJ, et al. The UCSC genome browser database: update 2010. Nucleic Acids Res. 2009;38:D613–D619. doi: 10.1093/nar/gkp939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.The UniProt Consortium. The Universal Protein Resource (UniProt) in 2010. Nucleic Acids Res. 2010;38:D142–D148. doi: 10.1093/nar/gkp846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Johnson AD, Handsaker RE, Pulit SL, Nizzari MM, O’Donnell CJ, de Bakker PI. SNAP: a web-based tool for identification and annotation of proxy SNPs using HapMap. Bioinformatics. 2008;24:2938–2939. doi: 10.1093/bioinformatics/btn564. [DOI] [PMC free article] [PubMed] [Google Scholar]