Abstract
We present the jump-start simultaneous alignment and tree construction using hidden Markov models (SATCHMO-JS) web server for simultaneous estimation of protein multiple sequence alignments (MSAs) and phylogenetic trees. The server takes as input a set of sequences in FASTA format, and outputs a phylogenetic tree and MSA; these can be viewed online or downloaded from the website. SATCHMO-JS is an extension of the SATCHMO algorithm, and employs a divide-and-conquer strategy to jump-start SATCHMO at a higher point in the phylogenetic tree, reducing the computational complexity of the progressive all-versus-all HMM–HMM scoring and alignment. Results on a benchmark dataset of 983 structurally aligned pairs from the PREFAB benchmark dataset show that SATCHMO-JS provides a statistically significant improvement in alignment accuracy over MUSCLE, Multiple Alignment using Fast Fourier Transform (MAFFT), ClustalW and the original SATCHMO algorithm. The SATCHMO-JS webserver is available at http://phylogenomics.berkeley.edu/satchmo-js. The datasets used in these experiments are available for download at http://phylogenomics.berkeley.edu/satchmo-js/supplementary/.
INTRODUCTION
Gene families develop novel functions and structures through evolutionary processes such as point mutation, insertion and deletion, gene duplication and domain architecture rearrangements (1). Shifts and increases in evolutionary rate (and development of novel functions) are common following gene duplication, resulting in neo- and sub-functionalization across paralogs. For these reasons, evolutionary reconstruction provides a natural framework for prediction of protein structure and function. Dobzhansky’s (2) famous saying, ‘Nothing in biology makes sense except in the light of evolution’ is literally true. Without evolution, interpreting biological data is almost impossible, and Dobzhansky’s proclamation might be accurately truncated to nothing in biology makes sense. However, bring in evolution and things start to fall into place; a picture starts to form. Structural phylogenomic analysis (3), in particular enables the detection of distant homologs and of correlations between changes in structure and changes in function. These (semi-orthogonal) structural and evolutionary analyses provide a computational and intellectual scaffold on which experimental and annotation data can be hung, allowing a nuanced view of the different structures and functions explored by a gene family—the molecular equivalent of Darwin’s endless forms most beautiful and most wonderful.
Evolutionary processes are central to the exploration of novel functions and structures; it follows that using phylogenetic information improves a host of bioinformatics methods including orthology prediction (4,5), functional site identification (6–8) and phylogenomic inference of gene function (9). For each of these tasks, the accuracy of the phylogenetic tree is essential.
The standard approach to phylogenetic reconstruction involves two separate steps: first, construct a multiple sequence alignment (MSA), and then estimate a phylogeny using the masked alignment as input. This two-step protocol is relatively straightforward when sequences in a dataset have high sequence identity (e.g. when sequences are restricted to orthologs from closely related species), since the MSA in these cases is likely to be accurate. However, alignment errors are common when datasets include remote homologs, with corresponding potential for error in the inferred phylogeny.
Errors in sequence alignment of distant homologs stem from two primary causes: sequence divergence causing reduced signal to guide the alignment, and actual changes in protein structure over evolution. While protein structure is generally conserved over large evolutionary distances, it is not totally immune to change. Numerous studies have shown that structural superposability drops with evolutionary distance; e.g. at <10% identity, only half of the residues may superpose (10). Accompanying the drop in structural superposability is an observed decrease in sequence alignment accuracy, assessed by agreement with a structural alignment. When two proteins have >30% identity, most alignment methods perform well, agreeing with the structural alignment for >90% of the structurally equivalent residue pairs. However, at <30% identity, accuracy starts to slide: at <20% identity, alignments can have quite serious errors, and at <15% identity only the best methods can get a small fraction of the structurally equivalent residues aligned correctly.
In general, simultaneous estimation of alignments and trees provides several advantages over the standard two-step process of phylogenetic tree estimation, independent of the specific techniques used. First, MSAs provide statements of positional homology, and are therefore an evolutionary hypothesis; separating the estimation of the tree from estimation of the alignment loses this valuable connection. Second, simultaneous estimation might avoid any systematic biases or errors encoded in an MSA by the alignment method selected. Third, simultaneous estimation can explore more of the alignment space, thus enabling greater accuracy. Fourth, a simultaneous estimation method can also make use of subtree-specific masking protocols [such as that used by simultaneous alignment and tree construction using hidden Markov models (SATCHMO)] to maximize the effective use of homology information in the dataset while avoiding the inclusion of non-homologous characters.
While several methods have been developed for co-estimation of phylogenies and MSAs for nucleotide data [e.g. SATé (11)], few are available for amino acid data. POY (12), a co-estimation method that seeks to minimize tree length, is able to handle both nucleotide and amino acid data and can take moderate-sized inputs, but has not been able to produce trees that are as accurate as the best two-phase methods (13) and our tests assessing POY on benchmark datasets of protein structural alignments do not show it to be competitive with the methods included in this article.
In addition to their use in phylogenetic tree estimation, MSAs are used in many bioinformatics tasks, including hidden Markov model (HMM) construction (14), predicting functional subfamilies (15), protein secondary structure prediction (16) and enzyme active site prediction (8).
SATCHMO and SATCHMO-JS
SATCHMO (17) employs progressive HMM–HMM scoring and alignment to estimate a phylogenetic tree and MSA. A novel subtree-specific masking procedure plays an important role in SATCHMO tree topology and alignment accuracy. During the SATCHMO progressive alignment and tree construction, pairs of subtrees are joined and new MSAs are created. SATCHMO analyzes each new MSA to identify regions that represent consensus structure across the sequences in that subtree; these are retained for HMM construction, and variable positions are forced into HMM insert states (represented by lowercase characters in the alignment). This subtree-specific masking protocol enables SATCHMO to take as input sequences sharing only a limited region of structural similarity (e.g. having one domain in common but different overall domain architectures). On the negative side, the SATCHMO algorithm, as originally defined, has two primary limitations: it is computationally very expensive, and the lack of iterative refinement means that errors introduced early in the process are not subsequently rectified.
SATCHMO-JS (jump-start SATCHMO) is an extension of the SATCHMO algorithm designed for scalability to large datasets. We use a divide-and-conquer approach to reduce complexity, employing the computationally efficient Multiple Alignment using Fast Fourier Transform (MAFFT) iterative MSA method (18) to align closely related sequences and saving the use of computationally expensive HMM–HMM alignment for only those subgroups that are more distantly related. Once a rooted tree is produced, Maximum Likelihood is used to optimize the tree edge lengths.
THE SATCHMO-JS WEB SERVER
Input
Users can submit up to 300 protein sequences in FASTA format at a time; the maximum allowable sequence length is 1000. Users supplying (optional) email addresses receive results by email; others can bookmark the results page. An Advanced Options page allows users to modify certain program defaults.
Processing method
The SATCHMO-JS pipeline starts with estimating an MSA for the input dataset using MAFFT (18). The MAFFT MSA is submitted to QuickTree (19) to estimate a Neighbor-Joining (NJ) tree. The MAFFT MSA and NJ tree are then analyzed to identify subtrees having a maximal allowable sequence divergence (program default is 35% identity; if a dataset is more closely related, we adjust this value accordingly). These subtree MSAs are submitted to the SATCHMO algorithm, jump-starting the method so that the HMM–HMM scoring and alignment only needs to be performed from that point ‘upwards’ to form a rooted tree and MSA. The tree topology within these subtrees is derived using the SCI-PHY algorithm (15). The SATCHMO tree and MSA are then submitted to RAxML (20) to optimize the tree edge lengths, keeping the SATCHMO tree topology fixed.
Following SATCHMO-JS progress
After sequence data has been submitted, the server forwards the user to a URL unique to the user’s submission. Until execution has completed, visiting this URL prompts the server to display a progress screen detailing the status of execution; results are displayed upon completion.
Output
Four primary results are returned. The first result is the SATCHMO MSA in University of California at Santa Cruz (UCSC) align-to-model (a2m) format. The UCSC a2m format is designed to display sequence paths through an HMM. Uppercase letters correspond to characters emitted in an HMM match state and represent the conserved structure across a dataset, while lowercase letters are emitted in an HMM insert state. Dashes correspond to paths through HMM delete (skip) states, and dots are inserted post hoc so that all sequences have the same number of characters in each column. The SATCHMO MSA can be viewed online or downloaded for examination using standard alignment viewers. Two SATCHMO trees—one with ML edge length optimization, and one with no edge lengths—are provided in Newick format. Lastly, we provide the SATCHMO tree-MSA (*.smo) file containing a novel data structure required for concurrent examination of the SATCHMO tree and MSA.
Web-based viewers
Users can view the SATCHMO tree and MSA using a modified version of the PhyloFacts Phyloscope viewer, as shown in Figure 1. Clicking on an internal node of the tree in the PhyloScope viewer will display the MSA for that subtree; MSAs near the root (which include more distant homologs) will have more lowercase characters (indicating these are not considered part of the conserved core for that subtree) than those near the leaves. MSA columns are colored according to conservation (based on BLOSUM62 scores). The tree and MSA can also be examined using Jalview (21).
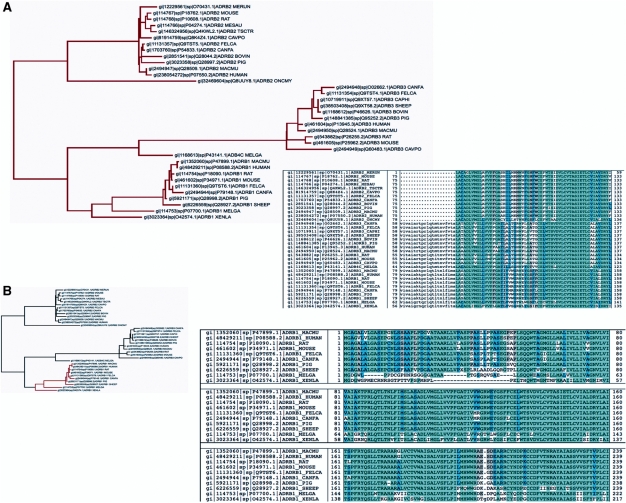
Figure 1.
Beta adrenergic receptors SATCHMO-JS tree and MSA displayed using the PhyloScope viewer. The PhyloScope viewer allows users to select internal nodes of the tree for examination of the alignments at these nodes, which may reflect different levels of inferred structural similarity across homologs. Columns are colored according to conservation based on BLOSUM62 sum-of-pairs scores (light blue indicates the highest level of conservation, followed by dark blue, grey and uncolored). Clicking on a subtree node restricts the MSA displayed to the sequences descending from that node, and highlights the selected subtree. The SATCHMO algorithm attempts to determine which columns are part of the conserved core structure across all sequences that descend from a node, resulting in some residues being displayed in lowercase (indicating that they are inserted relative to the consensus) at nodes higher in the tree (toward the root) but in uppercase at subtrees nearer the leaves. (A) The SATCHMO-JS tree and MSA corresponding to the root of the tree, where all sequences are selected. The first ∼70 residues of most sequences display in lowercase (indicating insertions relative to the consensus structure) reflecting structural variability over the dataset as a whole in this region. (Coincidentally, the region identified by SATCHMO as conserved across the dataset corresponds to the PFAM 7TM_1 HMM, which matches this region.) (B) The ADRB1 subtree (corresponding to orthologous Beta-1 adrenergic receptors from different species) has been selected by clicking the subtree node. This results in coloring the selected subtree red and displaying the MSA corresponding to sequences descending from that node. Note that many residues that displayed in lowercase in the SATCHMO root-level MSA are now displayed in uppercase, indicating that they are predicted by SATCHMO to be part of the conserve core structure for Beta-1 adrenergic receptors. Examining this subtree MSA shows that ADRB1_XENLA (from Xenopus laevis, African clawed frog) and ADRB1_MEGLA (from Meleagris gallopavo, Common turkey) diverge from mammalian orthologs at the N-terminus.
Downloads
The SATCHMO-JS tree, MSA and *.smo file can be downloaded from the results page.
BENCHMARKING SATCHMO-JS
We evaluated SATCHMO-JS performance on a dataset of 983 structurally aligned protein domains from the PREFAB benchmark dataset (22). We compared the original SATCHMO, SATCHMO-JS, MAFFT (v6.710b, five iterations refinement) (18), MUSCLE (v3.7, five iterations refinement) (22) and ClustalW (v2.0.12) (23).
PREFAB is composed of pairs of homologous structural domains that have been structurally superposed to obtain a reference alignment, and includes at most 50 homologs for each pair. This dataset is convenient for benchmarking alignment accuracy, but does not represent the typical data given as input to MSA programs. First, sequences in an MSA are rarely restricted to their mutually homologous domains (and structure information useful for this purpose is sparse). Second, protein MSAs are typically estimated using a larger number of (generally full-length) sequences. In addition, changes in overall domain architecture are quite common among sequences sharing significant sequence similarity. In these experiments, we sought to simulate the actual use of MSA methods in practice: we used each reference alignment as is, but modified the homolog selection protocol as follows. We retrieved the full-length protein for each structural domain and used it as a query to gather glocal (global–local, or semi-global, requiring sequences to match the query but allowing N- and C-terminal extensions) homologs from the UniRef90 dataset (a subset of UniProt such that no two sequences have >90% identity) using FlowerPower (24). Sequences retrieved were modified to remove any residues preceding and following the alignment to the full-length protein query, followed by removing indel characters (dots and dashes) to produce a set of FASTA (sub)sequences. The union of the two sets of (sub)sequences, including the reference domains, was given to each method to derive a MSA, followed by extracting the pairwise alignment of the two domains from the MSA. (In the case of SATCHMO, we evaluated the alignment corresponding to the first point in the tree at which the two sequences were joined in the SATCHMO tree.) The pairwise sequence alignment was then compared against the reference structural alignment using different scoring functions.
Results
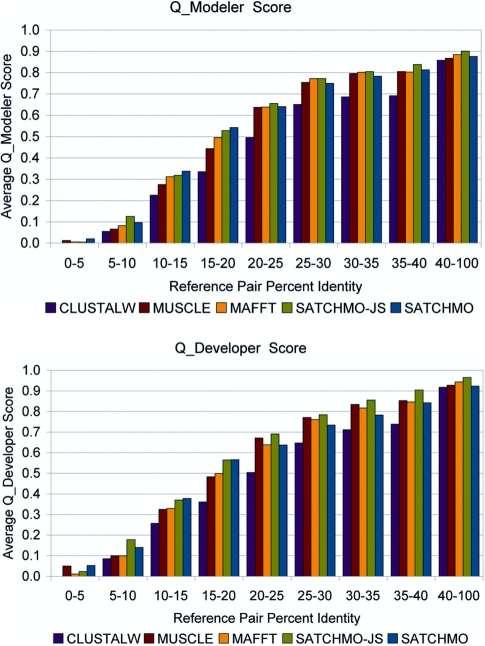
Figure 2 shows results on two scoring functions comparing the sequence alignment to the reference structural alignment. The Developer score (Qdeveloper) penalizes under-alignment relative to the reference, while the Modeler score (Qmodeler) penalizes over-alignment; these are thus measures of alignment recall and precision, respectively (25). As shown here, SATCHMO-JS produces alignments with greater concordance with the reference structural alignment than the original SATCHMO algorithm, MUSCLE, MAFFT, and ClustalW. Across the dataset as a whole, SATCHMO-JS provides a statistically significant improvement in Qdeveloper scores (Wilcoxon paired score signed rank test P < 0.05) relative to all methods. In Qmodeler scores, SATCHMO-JS provides a statistically significant improvement over all methods with the exception of MAFFT; here, SATCHMO-JS improves Qmodeler scores, but the difference is less pronounced (P = 0.204). Results for Qcombined and Cline Shift scores (26), which balance recall and precision, also show SATCHMO-JS to provide improved performance (see Supplementary Material).
Figure 2.
Benchmarking MSA accuracy. Methods used in this comparison include the original SATCHMO, SATCHMO-JS, ClustalW, MUSCLE and MAFFT (MUSCLE and MAFFT each used five iterations refinement). Results are shown on 983 pairs from the PREFAB benchmark dataset, divided into bins based on the percent identity in the reference structural alignment. The Modeler score (Qmodeler) is a measure of the precision of an alignment, while the Developer score (Qdeveloper) is a measure of the recall. For every percent identity bin, either SATCHMO or SATCHMO-JS produces the best overall performance in both Modeler and Developer scores, with SATCHMO-JS generally producing better results than SATCHMO. Over the dataset as a whole, SATCHMO-JS’s improvement relative to other methods tested is statistically significant (P < 0.05 using Wilcoxon paired score signed rank tests) for all scoring functions (including Qcombined and the Cline Shift score, which balance recall and precision) with a single exception: relative to MAFFT, the difference is significant only for the Developer score (P = 1.138e-05). For the Modeler, Qcombined and Cline Shift scores, the P-values are 0.204, 0.093 and 0.157, respectively. See text for additional details.
Figure 2 also explains our reasons for using MAFFT to align closely related sequences in the divide-and-conquer strategy used in SATCHMO-JS. Examining both the Modeler and Developer scores for different pairwise identity ranges shows that the original SATCHMO algorithm yields better results than MAFFT, MUSCLE and ClustalW (and sometimes also SATCHMO-JS) for very distant homologs having <20% identity, but that MUSCLE and MAFFT produce better results than the original SATCHMO above 25% identity. Our divide-and-conquer approach enables us to obtain the best of both approaches.
Datasets used in these experiments and additional results are available from http://phylogenomics.berkeley.edu/satchmo-js/supplementary/.
Computational efficiency
The jumpstart protocol allows SATCHMO-JS to handle large datasets. While the number of sequences in the input may be in the hundreds, the HMM–HMM scoring and alignment is restricted to perhaps a few dozen subtrees, reducing the run-time dramatically for most inputs. See Table 1.
Table 1.
Compute time required to estimate MSAs of different sizes, measured in seconds
| Size/length | SATCHMO-JS | SATCHMO | ProbCons | T-Coffee | MAFFT |
|---|---|---|---|---|---|
| 100/230 | 30.09 | 198.85 | 85.99 | 219.74 | 2.14 |
| 200/155 | 112.49 | 346.06 | 265.23 | 954.96 | 13.94 |
| 300/126 | 234.79 | 533.97 | 560.93 | 3882.01 | 23.84 |
| 500/392 | 1085.12 | 14 393.87 | 10 469.5 | — | 232.94 |
The first column gives the number of sequences and average sequence length for each dataset. ProbCons and MAFFT were run with five iterations of refinement; SATCHMO, SATCHMO-JS and T-Coffee used default parameters. The time to run SATCHMO-JS includes the time required for MAFFT, QuickTree and the subtree-selection program. MUSCLE’s run-time on these datasets is slightly longer than that of MAFFT (data not shown). T-Coffee failed to complete on the dataset with 500 sequences.
DISCUSSION
The SATCHMO-JS web server provides an extension of the SATCHMO algorithm for simultaneous MSA and tree construction for protein sequences. An interactive tree-MSA viewer allows users to examine MSAs at different points in a phylogenetic tree, and to download results for local viewing or for input to other programs.
SATCHMO-JS addresses certain limitations of the original SATCHMO algorithm through a divide-and-conquer approach: we partition an input dataset into smaller pieces that can be accurately aligned by a fast alignment method such as MAFFT, and then recombine these separate solutions into a whole using the computationally expensive SATCHMO HMM–HMM alignment. By restricting the computationally expensive HMM–HMM scoring and alignment steps to regions higher in the tree, we reduce the computational cost of the overall alignment task. At the same time, this process improves the overall accuracy, since the HMMs located at subtree nodes for subgroups identified by the divide-and-conquer protocol are based on more accurate MSAs. These HMMs provide a better basis for estimating the tree and MSA at higher regions in the tree (i.e. from that point upwards to the root).
The experimental design used here to compare SATCHMO-JS versus other MSA programs is intended to simulate the typical sequence selection process used in practice. These data, which may contain proteins with different overall domain architectures, present a significant challenge to alignment methods (many of which assume that input sequences are globally alignable). Since domain shuffling events and the presence of promiscuous domains can cause many homologs retrieved by BLAST and PSI-BLAST to have different overall domain architectures, methods that can handle these data appropriately are critical. Our results show that SATCHMO and SATCHMO-JS are more robust under these conditions than are MAFFT, MUSCLE and ClustalW. We expect that this improved performance is due to the use of subtree-specific masking during the SATCHMO hierarchical tree estimation, allowing SATCHMO to focus on regions that can be predicted to be structurally equivalent, facilitating local–local alignment.
FUTURE WORK
Due to their computational complexity, ProbCons (27) and T-Coffee (28) (as shown in Table 1) were not included in these experiments; comparisons versus these methods are planned for a future publication. We also plan additional extensions to the SATCHMO-JS algorithm. Web server modifications planned include additional options on the Advanced Settings page, extensions to the Phyloscope tree viewer functionality, and a Mac version of the SATCHMO MSA/tree viewer (to read *.smo files).
FUNDING
Funding: National Science Foundation (grant #0732065).
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We are grateful to Tandy Warnow for inspiring the use of the divide-and-conquer strategy used here, and to anonymous referees for helpful comments.
REFERENCES
- 1.Sjölander K. Phylogenomic inference of protein molecular function: advances and challenges. Bioinformatics. 2004;20:170–179. doi: 10.1093/bioinformatics/bth021. [DOI] [PubMed] [Google Scholar]
- 2.Dobzhansky CT. Nothing in biology makes sense except in the light of evolution. Am. Biol. Teach. 1973;35:125–129. [Google Scholar]
- 3.Sjölander K. Getting started in structural phylogenomics. PLoS Comput. Biol. 2010;6:e1000621. doi: 10.1371/journal.pcbi.1000621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Datta RS, Meacham C, Samad B, Neyer C, Sjölander K. Berkeley PHOG: PhyloFacts orthology group prediction web server. Nucleic Acids Res. 2009;37:W84–W89. doi: 10.1093/nar/gkp373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gabaldon T. Large-scale assignment of orthology: back to phylogenetics? Genome Biol. 2008;9:235. doi: 10.1186/gb-2008-9-10-235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sankararaman S, Sjölander K. INTREPID–INformation-theoretic TREe traversal for protein functional site IDentification. Bioinformatics. 2008;24:2445–2452. doi: 10.1093/bioinformatics/btn474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Capra JA, Laskowski RA, Thornton JM, Singh M, Funkhouser TA. Predicting protein ligand binding sites by combining evolutionary sequence conservation and 3D structure. PLoS Comput. Biol. 2009;5:e1000585. doi: 10.1371/journal.pcbi.1000585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sankararaman S, Sha F, Kirsch JF, Jordan MI, Sjölander K. Active site prediction using evolutionary and structural information. Bioinformatics. 2010;26:617–624. doi: 10.1093/bioinformatics/btq008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brown D, Sjölander K. Functional classification using phylogenomic inference. PLoS Comput. Biol. 2006;2:e77. doi: 10.1371/journal.pcbi.0020077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chothia C, Lesk AM. The relation between the divergence of sequence and structure in proteins. EMBO J. 1986;5:823–826. doi: 10.1002/j.1460-2075.1986.tb04288.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu K, Raghavan S, Nelesen S, Linder CR, Warnow T. Rapid and accurate large-scale coestimation of sequence alignments and phylogenetic trees. Science. 2009;324:1561–1564. doi: 10.1126/science.1171243. [DOI] [PubMed] [Google Scholar]
- 12.Varón A, Vinh LS, Wheeler WC. POY version 4: phylogenetic analysis using dynamic homologies. Cladistics. 2010;26:72–85. doi: 10.1111/j.1096-0031.2009.00282.x. [DOI] [PubMed] [Google Scholar]
- 13.Liu K, Nelesen S, Raghavan S, Linder CR, Warnow T. Barking up the wrong treelength: the impact of gap penalty on alignment and tree accuracy. IEEE/ACM Trans. Comput. Biol. Bioinform. 2009;6:7–21. doi: 10.1109/TCBB.2008.63. [DOI] [PubMed] [Google Scholar]
- 14.Krogh A, Brown M, Mian IS, Sjölander K, Haussler D. Hidden Markov models in computational biology. Applications to protein modeling. J. Mol. Biol. 1994;235:1501–1531. doi: 10.1006/jmbi.1994.1104. [DOI] [PubMed] [Google Scholar]
- 15.Brown DP, Krishnamurthy N, Sjölander K. Automated protein subfamily identification and classification. PLoS Comput. Biol. 2007;3:e160. doi: 10.1371/journal.pcbi.0030160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McGuffin LJ, Bryson K, Jones DT. The PSIPRED protein structure prediction server. Bioinformatics. 2000;16:404–405. doi: 10.1093/bioinformatics/16.4.404. [DOI] [PubMed] [Google Scholar]
- 17.Edgar RC, Sjölander K. SATCHMO: sequence alignment and tree construction using hidden Markov models. Bioinformatics. 2003;19:1404–1411. doi: 10.1093/bioinformatics/btg158. [DOI] [PubMed] [Google Scholar]
- 18.Katoh K, Toh H. Recent developments in the MAFFT multiple sequence alignment program. Brief. Bioinform. 2008;9:286–298. doi: 10.1093/bib/bbn013. [DOI] [PubMed] [Google Scholar]
- 19.Howe K, Bateman A, Durbin R. QuickTree: building huge Neighbour-Joining trees of protein sequences. Bioinformatics. 2002;18:1546–1547. doi: 10.1093/bioinformatics/18.11.1546. [DOI] [PubMed] [Google Scholar]
- 20.Stamatakis A, Ludwig T, Meier H. RAxML-III: a fast program for maximum likelihood-based inference of large phylogenetic trees. Bioinformatics. 2005;21:456–463. doi: 10.1093/bioinformatics/bti191. [DOI] [PubMed] [Google Scholar]
- 21.Waterhouse AM, Procter JB, Martin DM, Clamp M, Barton GJ. Jalview Version 2—a multiple sequence alignment editor and analysis workbench. Bioinformatics. 2009;25:1189–1191. doi: 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 24.Krishnamurthy N, Brown D, Sjölander K. FlowerPower: clustering proteins into domain architecture classes for phylogenomic inference of protein function. BMC Evol. Biol. 2007;7(Suppl. 1):S12. doi: 10.1186/1471-2148-7-S1-S12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang G, Dunbrack R.L., Jr Scoring profile-to-profile sequence alignments. Protein Sci. 2004;13:1612–1626. doi: 10.1110/ps.03601504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cline M, Hughey R, Karplus K. Predicting reliable regions in protein sequence alignments. Bioinformatics. 2002;18:306–314. doi: 10.1093/bioinformatics/18.2.306. [DOI] [PubMed] [Google Scholar]
- 27.Do CB, Mahabhashyam MSP, Brudno M, Batzoglou S. ProbCons: Probabilistic consistency-based multiple sequence alignment. Genome Res. 2005;15:330–340. doi: 10.1101/gr.2821705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Notredame C, Higgins DG, Heringa J. T-Coffee: a novel method for fast and accurate multiple sequence alignment. J. Mol. Biol. 2000;302:205–217. doi: 10.1006/jmbi.2000.4042. [DOI] [PubMed] [Google Scholar]