Abstract
Evaluating the accuracy of multiple sequence alignment (MSA) is critical for virtually every comparative sequence analysis that uses an MSA as input. Here we present the GUIDANCE web-server, a user-friendly, open access tool for the identification of unreliable alignment regions. The web-server accepts as input a set of unaligned sequences. The server aligns the sequences and provides a simple graphic visualization of the confidence score of each column, residue and sequence of an alignment, using a color-coding scheme. The method is generic and the user is allowed to choose the alignment algorithm (ClustalW, MAFFT and PRANK are supported) as well as any type of molecular sequences (nucleotide, protein or codon sequences). The server implements two different algorithms for evaluating confidence scores: (i) the heads-or-tails (HoT) method, which measures alignment uncertainty due to co-optimal solutions; (ii) the GUIDANCE method, which measures the robustness of the alignment to guide-tree uncertainty. The server projects the confidence scores onto the MSA and points to columns and sequences that are unreliably aligned. These can be automatically removed in preparation for downstream analyses. GUIDANCE is freely available for use at http://guidance.tau.ac.il.
INTRODUCTION
Multiple sequence alignment (MSA) is the foundation for most comparative sequence studies, including phylogeny reconstruction, characterization of functional protein sites, structural protein alignment and modeling, inference of selection forces and profile based homology search. The input MSA is taken for granted in most such studies, regardless of uncertainties in the alignment. However, typical sequence data sets usually result in MSAs harboring numerous errors, which are expected to affect the downstream analysis. Thus, researchers should acknowledge alignment uncertainty and take measures to account for and contain its effect on their analyses.
Here we present the GUIDANCE web-server, a tool for assigning a confidence score for each residue, column and sequence in an alignment, and for projecting these scores onto the MSA. The server points to columns and sequences that are unreliably aligned and enables their automatic removal from the MSA, in preparation for downstream analyses. The GUIDANCE server has a user-friendly interface, intuitive graphical results, and is freely available for use at http://guidance.tau.ac.il with no requirement of log-in.
Two algorithms for quantifying MSA uncertainties are implemented in the server. The GUIDANCE score is based on the robustness of the MSA to guide-tree uncertainty and relies on the bootstrap approach (1). The heads-or-tails (HoT) score measures alignment uncertainty due to co-optimal solutions (2,3).
Similar tools exist for assessing alignment confidence, such as T-COFFEE, SOAP and MUMSA (4–6). The advantages of the web-server implemented here are: (i) it is based on robust statistical measures of MSA reliability for quantifying two major sources of alignment uncertainty (co-optimal solutions and guide-tree uncertainty) that are not addressed by other tools; (ii) it allows the user to fine-tune the degree to which unreliable MSA parts are removed; (iii) it implements a range of MSA algorithms and evolutionary models (for codons, amino acids and nucleotides); and (iv) it is straightforward and easy to use.
The GUIDANCE and HoT methods were extensively tested and evaluated in several benchmark studies (1–3) that measured their predictive power for detecting alignment errors. These studies included both simulated protein sequence benchmarks, using the simulators ROSE (7) and INDELible (8), and the widely-accepted real biological benchmark BAliBASE (9). Both methods score very high in receiver operating characteristics (ROC) analysis that measures their sensitivity and specificity as predictors for alignment errors. The combined predictive power measured in terms of the area under the ROC curve (AUC) on the BAliBASE benchmark was 94.0 and 89.7% for GUIDANCE and HoT, respectively, and on the simulations benchmark—96.5 and 92.8% (1). Therefore, alignment regions marked unreliable by the GUIDANCE server are expected to accurately correlate with the large majority of actual alignment errors.
METHODS
The GUIDANCE web server implements the HoT algorithm (2) and the GUIDANCE algorithm (1) for the assessment of alignment uncertainty. The minimal input requirement for both algorithms is: (i) DNA, RNA or protein sequences in FASTA format; and (ii) a choice of MSA algorithm. Using these two inputs the following steps are carried out: (i) a standard MSA is generated, hereby termed ‘base MSA’, by applying the MSA algorithm; (ii) a set of perturbed MSAs is constructed according to the alignment confidence algorithm (HoT of GUIDANCE, see below); (iii) the set of MSAs is compared to the base MSA in order to estimate its confidence level. This comparison results in confidence scores between 0 and 1 for each residue, residue pair, column and sequence of the MSA, which are essentially different ways to average sum-of-pairs (SP) scores (10,11); (iv) the confidence scores of all residues are projected onto the MSA, using a color-scale and the column scores are plotted below the alignment; and (v) unreliable columns and sequences may be removed from the base MSA. The server currently supports three progressive alignment algorithms: ClustalW, MAFFT and PRANK (12–14).
The above procedure differs between GUIDANCE and HoT in the way that the set of perturbed MSA is created. GUIDANCE scores reflect the robustness of an alignment to guide tree uncertainty. The GUIDANCE method perturbs the guide tree used to build the MSA, using bootstrap sampling (1). On the other hand, HoT scores reflect alignment uncertainty due to co-optimal solutions in the progressive alignment procedure. Here the set of perturbed MSAs is constructed by reversing the sequences at each of the pairwise alignment steps of the progressive alignment algorithm (2).
Adjustable parameters
The server implements a few advanced options that are useful for fine-tuning the results. For the GUIDANCE algorithm, the number of bootstrap repeats can be set by the user (the default value is set to 100). The higher this number is, the more accurate the confidence score, but the running time increases linearly. The cutoffs according to which columns and sequences are filtered out for subsequent analysis are also adjustable. It is possible to change these cutoffs according to the proportion of columns\sequences that the user wishes to retain. The order of the sequences in the output MSA may be set according to the input file, or according to the alignment algorithm result file.
In addition, the server allows uploading a user MSA file instead of the sequences file. In this case, the input MSA is used as the base MSA and the confidence scores are calculated in the same way as described above. This option should be used with caution. It is useful for analyzing an MSA of interest, for example, an MSA that was generated using a more accurate guide-tree than the standard neighbor joining tree. However, it is important to remember that even when the base MSA is given as input, the alignment algorithm chosen is applied many times in order to generate each of the perturbed MSAs. Therefore, supplying an MSA created by one program and inferring its confidence using another program may result in false predictions.
Advanced users can also alter the parameters passed on to the alignment program used. For example, by default, the server runs PRANK with the ‘+F’ flag, but the experienced user may wish to remove that option in some cases (http://www.ebi.ac.uk/goldman-srv/prank/). For MAFFT the user may enable the iterative refinement option and set the number of iterations in the MAXITERATE parameter. Additionally, an option to choose between the iterative refinement strategies genafpair, localpair and globalpair is provided when running MAFFT. See the MAFFT website for a description of these options (http://mafft.cbrc.jp/alignment/software/algorithms/algorithms.html).
Output
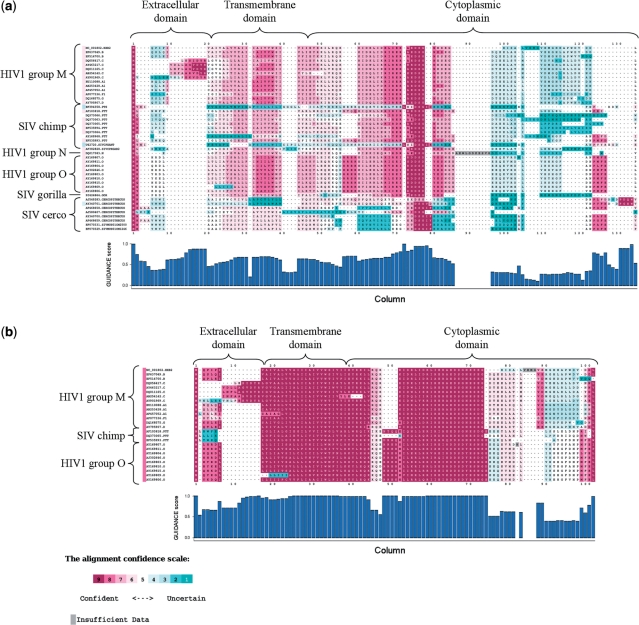
The main result of the GUIDANCE server is a graphical visualization of the confidence scores which consists of two parts (Figure 1a): (i) color-scaled projection of the confidence scores of each residue onto the base MSA; and (ii) a plot of the column scores. Text files are also produced containing the confidence scores for each column, residue, residue-pair and sequence. In addition, the following MSA files are provided: (i) the base MSA; (ii) the MSA containing only reliable columns that passed a predefined threshold (this file may be used in downstream analyses such as phylogeny reconstruction); and (iii) a sequence file of reliable sequences only (again for a pre-defined threshold). It is recommended to rerun GUIDANCE on the filtered sequences as input, in order to re-align them without the disruptive effect of the badly aligned sequences (see example below). This can be done simply by clicking on the button next to the output file link.
Figure 1.
An example of the GUIDANCE output. (a) Residue confidence scores are projected onto the MAFFT alignment of Vpu protein sequences from human and simian immunodeficiency viruses (HIV and SIV). Confidently aligned residues are colored in shades of magenta and pink, while uncertain residues are colored in shades of blue. Column scores are plotted below the alignment. (b) Dramatically improved alignment confidence after filtering low-scoring sequences and re-running GUIDANCE. Note the color-coding next to the sequence names before and after re-alignment.
Implementation
The GUIDANCE web server runs on a Linux cluster of 2.6 GHz AMD Opteron processors, equipped with 4 GB RAM per quad-core node. At the moment, our cluster will allocate up to 16 cores for GUIDANCE runs submitted through the web server, and the allocation of resources will grow with demand. The server runs up-to-date versions of the supported multiple alignment programs and an in-house implementation of neighbor joining bootstrap tree reconstruction. The HoT and GUIDANCE algorithms are implemented in Perl and C++. The source code of GUIDANCE is also available on the website, for large scale analyses, which users may want to run locally using their own computational resources.
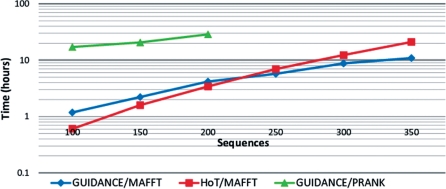
Running time depends on the data set size (number and length of sequences) and (for GUIDANCE scores) on the number of bootstrap repeats. The major component of the running time is the multiple alignment program used, thus MAFFT runs will be fastest and PRANK runs slowest. To aid users with estimating running time for their data sets, we include a plot of average GUIDANCE and HoT running times using either MAFFT or PRANK for several data set sizes, from 100 to 350 sequences, roughly 300 amino acids in length (Figure 2). Note that GUIDANCE was run with the default 100 bootstrap repeats, but this number can be reduced to shorten the running time. HoT running time depends on the number of branches in the guide tree, which increases linearly with the number of sequences.
Figure 2.
Average run-time performance as a function of the number of sequences. Simulated protein sequences roughly 300 amino acids long were aligned using MAFFT and analyzed by GUIDANCE (blue diamonds) or HoT (red squares). In addition, running time for GUIDANCE on PRANK alignments is plotted with green triangles. Each data point represents ten replicates.
Recommended usage
GUIDANCE is recommended for use in conjunction with any and all MSA-based studies, since virtually all MSAs are affected by some degree of uncertainty. The type of downstream analysis may dictate different modes of running GUIDANCE and of usage of GUIDANCE scores. It is generally recommended to use GUIDANCE to filter out badly aligned sequences and re-align the data, since such sequences usually disrupt the alignment among the other sequences, which could be reliably aligned otherwise. However, the option for removing alignment columns may or may not be used, depending on the expected sensitivity of the analysis to alignment errors.
For example, site-specific rate inference, as in the Consurf web server (15), is usually robust to a few badly aligned residues in a column, because a column corresponding to a conserved site will still be inferred as conserved as long as most of the data are correctly aligned. Conversely, site-specific prediction of positive selection using the Ka/Ks measure, as in the Selecton web server (16), may be sensitive to a few badly aligned residues that can inflate the Ka/Ks estimate for the column and lead to false inference of positive selection (17). Moreover, Ka/Ks inference of positive selection is only considered if the whole gene passes an LRT statistical significance threshold. Therefore, the inclusion of badly aligned columns in this test may be detrimental for certain genes that erroneously pass the LRT threshold due to the inflated Ka/Ks scores in these columns.
Perhaps the most widely used MSA-based analysis is phylogeny reconstruction. It is common practice to filter gap-less blocks in the alignment and only use those columns for phylogeny reconstruction. GBLOCKS (18,19) is usually used for this purpose. A comparative study has demonstrated that the accuracy of filtering columns containing alignment errors by GUIDANCE is superior over GBLOCKS (1). The merits of removing columns for phylogeny reconstruction may vary between different data sets and different evolutionary scenarios because of the delicate balance between filtering noise and loss of evolutionary information. Therefore, it is debated whether columns should be removed for phylogeny reconstruction (e.g. 20–22).
Furthermore, the choice of cutoff on the confidence scores clearly affects the tradeoff between the sensitivity and the specificity in the identification of alignment errors. There are no specific recommended values for these cutoffs because their effect on the alignment varies considerably among data sets. The web server provides a list of cutoffs with their respective effects on the remaining proportion of sequences/columns and users are encouraged to experiment with several cutoffs, especially when removing sequences and re-aligning the data set.
In general, it is recommended to use the GUIDANCE method, as it was demonstrated to outperform HoT on both the BAliBASE benchmark and on simulations studies (1). However, in a few cases the guide tree may be highly robust, which may lead to an overestimation of the confidence scores produced by GUIDANCE, or there may be a single guide tree, such as in the alignment of two or three sequences. In such cases, the GUIDANCE scores will be uninformative, and HoT, which is not affected by the guide tree, should be used.
CASE STUDY: THE HIV Vpu ACCESSORY PROTEIN
We illustrate using GUIDANCE to identify unreliable alignment columns by analyzing an MSA of Vpu protein sequences from human and simian immunodeficiency viruses (HIV and SIV). These viruses are known for their high rate of evolution, which is attributed to an arms race between the virus and the host immune system (23). The Vpu protein has been recently shown to antagonize the host protein Tetherin, an innate immune factor, in order to promote viral release and replication (24). Therefore, it is a natural candidate for many evolutionary analyses that rely on an MSA. Our purpose here was to demonstrate the use of GUIDANCE to evaluate reliability of the MSA, and the importance of this evaluation for the interpretation of downstream analyses.
Vpu is an accessory viral protein present in HIV-1 and SIV infecting chimpanzees and other primate species, yet absent in HIV-2. The protein contains ∼80 amino acids and has two known major functions, which are conducted by two distinct domains of the protein: (i) promotion of CD4 degradation via the cytoplasmic domain; and (ii) enhancement of virion release from host cells via the transmembrane domain, which was implicated in antagonism of Tetherin (24,25).
We ran GUIDANCE on a sample of Vpu protein sequences from the three main HIV-1 groups (M, N and O) and SIV sequences from chimpanzee (Pan troglodytes), gorilla (Gorilla gorilla) and several Cercopithecus species, using MAFFT. The results clearly show that the alignment of the cytoplasmic domain of Vpu is not robust to perturbations in the guide tree. The same applies to some residues in the transmembrane and extracellular domains (Figure 1a). Looking at specific sequences, the SIV sequences from Cercopithecus and some of the sequences from P. troglodytes are shown to be badly aligned with the rest of the sequence set. By simply pressing a button, these sequences were filtered and GUIDANCE was rerun on the confidently aligned sequences only. The results demonstrate a dramatic improvement in MSA confidence (Figure 1b). The transmembrane domain and the 5′ region of the cytoplasmic domain now receive almost perfect confidence scores. Note that although a clade of sequences was excluded by the GUIDANCE filter and the alignment is now considerably more condense, the remaining sequences are still highly variable and several gapped regions have been retained. The removal of the unconfidently-aligned sequences is necessary to avoid artifacts that they would have otherwise caused in downstream analyses such as inference of positive selection (17,26).
Even after removing the low-scoring sequences, the alignment of the 3′ region of the cytoplasmic domain is uncertain, thus, downstream analyses on these regions should be interpreted with caution. When appropriate, one may use the filtered MSA, provided by GUIDANCE, which contains only the reliable columns (e.g. for inference of positive selection). This example demonstrates the importance of using GUIDANCE for removing badly aligned sequences that may disrupt the MSA and for noting which columns are suspect of alignment errors, which might affect downstream analysis.
FUNDING
Israel Science Foundation (grant 878/09 to T.P.); Saia foundation (HIV research to T.P.); US National Library of Medicine (grant LM010009-01 to D.G. and G.L.); Converging Technologies Program (to O.P.); and Edmond J. Safra Program (E.P.). Funding for open access charge: Israel Science Foundation (grant 878/09).
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We thank the three anonymous reviewers for their constructive comments.
REFERENCES
- 1.Penn O., Privman E., Landan G., Graur D., Pupko T. An alignment confidence score capturing robustness to guide-tree uncertainty. Mol. Biol. Evol. 2010 doi: 10.1093/molbev/msq066. doi:10.1093/molbev/msq066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Landan G., Graur D. Local reliability measures from sets of co-optimal multiple sequence alignments. Pacific Symp. Biocomput. 2008;13:15–24. [PubMed] [Google Scholar]
- 3.Landan G., Graur D. Heads or tails: a simple reliability check for multiple sequence alignments. Mol. Biol. Evol. 2007;24:1380–1383. doi: 10.1093/molbev/msm060. [DOI] [PubMed] [Google Scholar]
- 4.Poirot O., O’Toole E., Notredame C. Tcoffee@igs: a web server for computing, evaluating and combining multiple sequence alignments. Nucleic Acids Res. 2003;31:3503–3506. doi: 10.1093/nar/gkg522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Loytynoja A., Milinkovitch M.C. SOAP, cleaning multiple alignments from unstable blocks. Bioinformatics. 2001;17:573–574. doi: 10.1093/bioinformatics/17.6.573. [DOI] [PubMed] [Google Scholar]
- 6.Lassmann T., Sonnhammer E.L. Automatic assessment of alignment quality. Nucleic Acids Res. 2005;33:7120–7128. doi: 10.1093/nar/gki1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stoye J., Evers D., Meyer F. Rose: generating sequence families. Bioinformatics. 1998;14:157–163. doi: 10.1093/bioinformatics/14.2.157. [DOI] [PubMed] [Google Scholar]
- 8.Fletcher W., Yang Z. INDELible: a flexible simulator of biological sequence evolution. Mol. Biol. Evol. 2009;26:1879–1888. doi: 10.1093/molbev/msp098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Thompson J.D., Koehl P., Ripp R., Poch O. BAliBASE 3.0: latest developments of the multiple sequence alignment benchmark. Proteins. 2005;61:127–136. doi: 10.1002/prot.20527. [DOI] [PubMed] [Google Scholar]
- 10.Thompson J.D., Plewniak F., Poch O. A comprehensive comparison of multiple sequence alignment programs. Nucleic Acids Res. 1999;27:2682–2690. doi: 10.1093/nar/27.13.2682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carrillo H., Lipman D. The multiple sequence alignment problem in biology. SIAM J. Appl. Math. 1988;48:1073–1082. [Google Scholar]
- 12.Katoh K., Kuma K., Toh H., Miyata T. MAFFT version 5: improvement in accuracy of multiple sequence alignment. Nucleic Acids Res. 2005;33:511–518. doi: 10.1093/nar/gki198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Thompson J.D., Higgins D.G., Gibson T.J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Loytynoja A., Goldman N. An algorithm for progressive multiple alignment of sequences with insertions. Proc. Natl Acad. Sci. USA. 2005;102:10557–10562. doi: 10.1073/pnas.0409137102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Landau M., Mayrose I., Rosenberg Y., Glaser F., Martz E., Pupko T., Ben-Tal N. ConSurf 2005: the projection of evolutionary conservation scores of residues on protein structures. Nucleic Acids Res. 2005;33:W299–W302. doi: 10.1093/nar/gki370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stern A., Doron-Faigenboim A., Erez E., Martz E., Bacharach E., Pupko T. Selecton 2007: advanced models for detecting positive and purifying selection using a Bayesian inference approach. Nucleic Acids Res. 2007;35:W506–W511. doi: 10.1093/nar/gkm382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schneider A., Souvorov A., Sabath N., Landan G., Gonnet G.H., Graur D. Estimates of positive Darwinian selection are inflated by errors in sequencing, annotation, and alignment. Genome Biol. Evol. 2009;2009:114. doi: 10.1093/gbe/evp012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Castresana J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol. Biol. Evol. 2000;17:540–552. doi: 10.1093/oxfordjournals.molbev.a026334. [DOI] [PubMed] [Google Scholar]
- 19.Talavera G., Castresana J. Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst. Biol. 2007;56:564–577. doi: 10.1080/10635150701472164. [DOI] [PubMed] [Google Scholar]
- 20.Gatesy J., DeSalle R., Wheeler W. Alignment-ambiguous nucleotide sites and the exclusion of systematic data. Mol. Phylogenet. Evol. 1993;2:152–157. doi: 10.1006/mpev.1993.1015. [DOI] [PubMed] [Google Scholar]
- 21.Giribet G., Wheeler W.C. On gaps. Mol. Phylogenet. Evol. 1999;13:132–143. doi: 10.1006/mpev.1999.0643. [DOI] [PubMed] [Google Scholar]
- 22.Aagesen L. The information content of an ambiguously alignable region, a case study of the trnL intron from the Rhamnaceae. Mol. Phylogenet. Evol. 2004;4:35–49. [Google Scholar]
- 23.Rambaut A., Posada D., Crandall K.A., Holmes E.C. The causes and consequences of HIV evolution. Nat. Rev. Genet. 2004;5:52–61. doi: 10.1038/nrg1246. [DOI] [PubMed] [Google Scholar]
- 24.Neil S.J., Zang T., Bieniasz P.D. Tetherin inhibits retrovirus release and is antagonized by HIV-1 Vpu. Nature. 2008;451:425–430. doi: 10.1038/nature06553. [DOI] [PubMed] [Google Scholar]
- 25.Nomaguchi M., Fujita M., Adachi A. Role of HIV-1 Vpu protein for virus spread and pathogenesis. Microbes Infect. 2008;10:960–967. doi: 10.1016/j.micinf.2008.07.006. [DOI] [PubMed] [Google Scholar]
- 26.Wong K.M., Suchard M.A., Huelsenbeck J.P. Alignment uncertainty and genomic analysis. Science. 2008;319:473–476. doi: 10.1126/science.1151532. [DOI] [PubMed] [Google Scholar]