Abstract
Harmonized typing of bacteria and easy identification of locally or internationally circulating clones are essential for epidemiological surveillance and disease control. For Mycobacterium tuberculosis complex (MTBC) species, multi-locus variable number tandem repeat analysis (MLVA) targeting mycobacterial interspersed repetitive units (MIRU) has been internationally adopted as the new standard, portable, reproducible and discriminatory typing method. However, no specialized bioinformatics web tools are available for analysing MLVA data in combination with other, complementary typing data. Therefore, we have developed the web application MIRU-VNTRplus (http://www.miru-vntrplus.org). This freely accessible service allows users to analyse genotyping data of their strains alone or in comparison with a reference database of strains representing the major MTBC lineages. Analysis and comparisons of genotypes can be based on MLVA-, spoligotype-, large sequence polymorphism and single nucleotide polymorphism data, or on a weighted combination of these markers. Tools for data exploration include search for similar strains, creation of phylogenetic and minimum spanning trees and mapping of geographic information. To facilitate scientific communication, an expanding genotype nomenclature (MLVA MtbC15-9 type) that can be queried via a web- or a SOAP-interface has been implemented. An extensive documentation guides users through all application functions.
INTRODUCTION
The bacteria of the Mycobacterium tuberculosis complex (MTBC) are the causative agents of tuberculosis (TB). This infectious disease is responsible for approximately 2 million deaths annually and foci of multi- and extensive drug resistance are emerging worldwide (1). Genotyping of MTBC strains empowers epidemiological surveillance and control, e.g. by permitting detection of clonal spread of multi-drug resistant strains and unsuspected outbreaks. At the clinical level, molecular typing enables identification of false positive cases due to laboratory cross-contamination and distinction between exogenous reinfection and relapse from initial infection. From a research perspective, molecular strain typing is valuable for the deciphering of the MTBC population structure and thus enabling the development of new diagnostics, vaccines or treatments (2).
The previous gold standard for MTBC genotyping—IS6110 restriction fragment length polymorphism (RFLP) typing—is laborious and time consuming and the resulting complex banding patterns make inter-laboratory comparisons difficult (3). As the MTBC gene sequence variation is low classical sequence-based typing methods such as multi-locus sequence typing, even extended to tens of genes, are only informative for the identification at genetic (sub-)lineage but not at strain level (4,5). Likewise, large sequence polymorphisms (LSP) or single nucleotide polymorphisms (SNP) elucidated MTBC genetic lineages differing in their geographical distribution immunogenicity, virulence and association with multidrug resistant TB (6–8).
However, the use of other, more variable markers is needed for a desirable finer phylogenetic classification and detection down to strain level to the advantage of public health and clinical objectives. As a portable, reproducible and discriminatory typing tool, the multi-locus variable number tandem repeat (VNTR) analysis (MLVA) targeting a number of tandem repeat loci including the mycobacterial interspersed repetitive units (MIRU) has, therefore, been internationally adopted as the new standard (9,10). Spoligotyping is often used as a rapid additional typing method to increase the discriminatory power of MIRU-VNTR typing. This method addresses the presence or absence of 43 spacers at the so-called direct repeat locus, a paradigmatic member of the family of clustered regularly interspaced short palindromic repeats (CRISPR) (11,12). In addition to their combined discriminatory power, 24-locus-based MIRU-VNTR typing and spoligotyping data are predictive of the main MTBC phylogenetic lineages, although they are somewhat less deterministic for such purposes than are LSP or sequence-based data (13,14).
Several web services containing strain collections of MLVA or spoligotyping data are available, e.g. MLVAbank or spolTools and SITVIT/SpolDB4 (15–18). Each of these web applications offers functions for simple data comparison and basic clustering for only one genotyping method. However, there was no online tool available that provided a comprehensive analysis of MTBC strains based on multiple genotyping methods and tools for directly naming new genotypes. We described recently such a system, i.e. MIRU-VNTRplus, with a focus on evaluation of a reference database that allows a robust-lineage identification based on the combination of different genotyping data (19). Here, we describe for the first time the complete functionality of MIRU-VNTRplus, including the new features minimum spanning tree (MST), geographic mapping and the nomenclature service.
IMPLEMENTATION
The MIRU-VNTRplus server was implemented in Java using the JavaServer Faces technology together with the RichFaces AJAX extension for creating the web pages. Java Servlets were used for image rendering. In order to store the data of the provided nomenclature service persistently, a MySQL database was used. Database access was implemented by means of the object-relational mapping framework Hibernate, while the SOAP interface for accessing the nomenclature service was realized by means of the Apache Axis2 framework. The mapping function uses Google Maps for geocoding and display of an interactive map. MIRU-VNTRplus is compatible with the most common web browsers, i.e. Mozilla Firefox 2 and 3; Microsoft Internet Explorer 6, 7 and 8; Opera 9 and 10; Apple Safari 4; and Google Chrome 3. To enable the application’s full functional capability JavaScript has to be activated.
The application allows the input of copy numbers for 24 MIRU-VNTR loci and 15 or 12 loci subsets (10). Furthermore, the following typing data are supported: presence or absence of 43 spoligotyping spacers, presence or absence of 15 standard LSPs (6,7), values of six SNPs (20) and susceptibility data for 16 drugs. Finally, in addition to general descriptive information (e.g. strain ID and country of isolation) input for three user-specific data fields can be supplied. Furthermore, a reference database containing gentoyping data of 186 strains representing the major MTBC lineages is available (19). IS6110 RFLP fingerprinting images are also shown for visual comparison of these strains.
Distance measures that can be used for strain comparison include categorical distance, DC, (δμ)2 and DSW (21–23). Double and variant alleles are regarded as own categories for categorical distance calculation and are treated as missing data for all other distance measures. When used for pairwise strain comparisons, the categorical distance is identical to the distance measure DA that is well suited for phylogenetic analysis of VNTR markers (24). Polyphasic typing is achieved by calculating combined pairwise distances for all chosen typing methods. A combined distance is calculated by multiplying the distance of each method by a weighting factor, summing up the distances for all methods, and dividing this value by the sum of the weighting factors. Missing data is ignored for the pairwise distance calculation, thus allowing users to work with a self-defined subset of loci.
Phylogenetic trees are inferred using the un-weighted pair group method with arithmetic means (UPGMA) or neighbour-joining (NJ) algorithms (25,26). Generated trees can be re-rooted using a manually selected outgroup. The resulting trees can be exported to Newick and NEXUS format and the underlying distance matrix can be downloaded as a MEGA file. A minimum spanning tree is created using Kruskal’s algorithm and a force-directed graph layout for visualization (27). All trees can be downloaded as raster image (PNG), vector image (SVG, EMF) or PDF document.
RESULTS
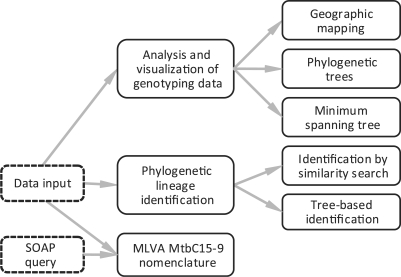
The freely accessible MIRU-VNTRplus web application (http://www.miru-vntrplus.org) offers three main functions: (i) phylogenetic lineage identification by using a reference database; (ii) analysis and visualization of genotyping data; and (iii) access to the MLVA MtbC15-9 nomenclature service (Figure 1). An extensive documentation including a manual, multi-media tutorials and protocols for the genotyping methods complements the web server.
Figure 1.
The three main functions of MIRU-VNTRplus are (i) phylogenetic lineage identification by using a reference database; (ii) analysis and visualization of genotyping data; and (iii) access to the MLVA MtbC15-9 nomenclature service.
Data input
Genotyping data can be entered for a single strain via a web form, copied from a spreadsheet application via clipboard or uploaded by using Comma Separated Values (CSV) or Microsoft (MS) Excel files. Template CSV and MS Excel files that simplify the upload process are available. MIRU-VNTRplus allows users to upload data for up to 500 strains. Various formats for genotyping data are accepted, e.g. MIRU-VNTR alleles with incomplete tandem repeat units (variant alleles) and two alleles simultaneously detected for a given locus (double alleles), CDC notation for MIRU-VNTR data (by using ‘A’ for 10 repeats, ‘B’ for 11 repeats, etc. double digits are avoided) or binary and octal numbers for spoligotyping patterns. After exclusion of possible PCR artefacts (i.e. classical stutter peaks) concordantly observed double alleles in several independent VNTR loci for a given sample indicate the presence of a mixed DNA population. This mixed population can result from a true mixed infection, or from culture or DNA contamination. In contrast, the occurrence of double alleles in a single locus rather suggests the presence of a given allelic variant within a clonal isolate. In the following, the entered or uploaded strain data is referred to as ‘user strains’.
Analysis and visualization of genotyping data
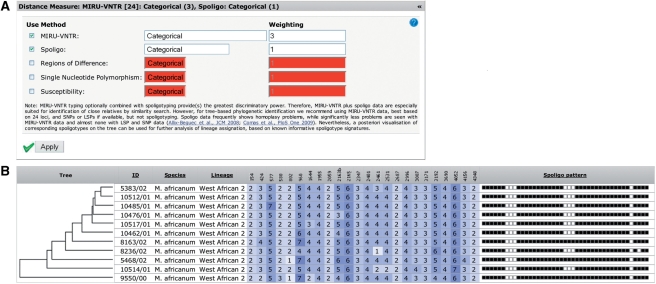
MIRU-VNTRplus supports polyphasic typing by calculating a combined distance for different types of genotyping data. An input form allows the selection of the used genotyping methods, distance measures and weightings (Figure 2A). This form can be accessed on all pages that use calculated distances. A data table that can be sorted by any column displays the strain data. Using the UPGMA or NJ algorithm a dendrogram can be directly included in this table, thus ordering the strains by their position in the tree (Figure 2B). A filter function allows one to exclude strains from analysis that do not match certain criteria. Complex filtering criteria can be created by combining comparison operators for certain data fields with the logical operators AND or OR. Strains can be marked manually or automatically with a background colour, e.g. according to the value of lineage, genetic marker or user data. By selecting a strain as the comparison strain all genetic marker differences are highlighted for all other strains. In addition the distances between the comparison strain and all other strains are displayed for sorting and filtering.
Figure 2.
(A) Input form allowing the selection of the used genotyping methods, distance measures and weightings. (B) Data exploration table. The data for each strain is displayed in a single row. Next to the ID, species and lineage columns, the table contains MIRU-VNTR data highlighted in blue shades, indicating the different copy numbers and spoligotyping patterns. An UPGMA tree is used for ordering the strains.
Phylogenetic trees can be created and modified interactively on an extra page. Depending on the user selection, trees are displayed as dendrograms or radial graphs. The label text can be chosen and genotyping data can be added to the image. It is possible to export the distance matrix, the tree and the resulting image to various formats. Clicking a branch or leaf of the tree opens a pop-up window that shows the genotyping data for all sub-tree strains and offers to re-root the tree, swap branches and mark sub-tree strains by colour as interactive features. In addition to classical phylogenetic trees, MIRU-VNTRplus also allows the calculation of MSTs. However, MSTs can be drawn based on data from only one typing method at a time. Clonal complexes (CC) as defined by genotypes sharing a selectable maximum locus difference are highlighted in the MST image. The choice of the label text, length of connection lines and zoom factor modifies the appearance of the MST. Using the context menu strains can again be marked by colour.
The strain-mapping feature displays the geographical distribution of strains on a map. The location information is retrieved from the data field country of isolation that might as well include ZIP codes or city information. The distribution of species, lineages or user data field values for each location is visualized in a colour-coded pie chart.
Phylogenetic lineage identification
The comparison with the reference database allows the identification of the phylogenetic lineage of user strains. Again, the identification can be based on a combined distance of several genotyping methods. Identification by similarity search lists reference database strains displaying an adjustable maximum distance to each user strain. The default maximum distance has been determined on the basis of validation tests using an external strain data set (19). The similarity search results can be used for the automatic assignment of species and lineage information to user strains. All results are exportable as MS Excel or CSV file. In most cases, identification by similarity search will not be sufficient to undoubtedly identify all user strains. Here, an additional tree-based analysis can be carried out by calculating a UPGMA or NJ tree that contains all user and reference database strains. Investigating strains that are monophyletic (i.e. ingroups) compared to reference database strains in the different tree branches allow the user to infer species and lineage assignment. Such assignments may then be set by clicking strains or nodes on the tree (Figure 3).
Figure 3.
Tree-based identification. Clicking a branch or leaf of the tree opens a popup window that offers functions for re-rooting the tree, swapping of branches and marking subtree strains by colour. Furthermore, species/lineage may be assigned for user strains (labelled with yellow boxes).
MtbC15-9 nomenclature service
For the exchange of MIRU-VNTR data, the reporting of the full genotype with copy numbers of each locus in perfect order is mandatory. To facilitate scientific communication, MIRU-VNTRplus introduces an expanding nomenclature that assigns a numerical code to MIRU-VNTR patterns. The MLVA MtbC15-9 type is a juxtaposition of two subtypes, i.e. the MtbC15 and MtbC9 type. These types are based on a set of the 15 most discriminatory MIRU-VNTR loci and a set of nine ancillary loci, as inferred by the analysis of single locus variation frequencies on a large international strain collection (10). The web application provides forms for converting MtbC15-9 types into VNTR copy numbers and vice versa either in single or batch mode. The results of these queries can be exported as CSV or MS Excel files. When uploading user strains MIRU-VNTRplus automatically detects and displays known MtbC15-9 types. New MtbC15-9 types can be added to the nomenclature service provided that contact information is supplied. An email that contains a link for confirmation of the assigned types is sent to the given contact email address. On the condition of confirmation, the new MtbC15-9 types are added to the nomenclature server and made publicly available. External applications may query the nomenclature service via a SOAP interface. The loci used for the MtbC15 and MtbC9 nomenclature, all existing types, the copy numbers for specific MtbC15 or MtbC9 types and the types for a given MIRU-VNTR pattern can be retrieved by SOAP queries. A documentation of the SOAP service that includes five examples in three different programming languages (Java, Perl and C#) is available for developers.
APPLICATION EXAMPLE
To provide an application case, a recently published data set containing MIRU-VNTR data and spoligotyping patterns for 97 MTBC strains has been re-analysed (28). These data can be loaded from the homepage of MIRU-VNTRplus by clicking the ‘open example dataset’ link. To reproduce the published results only the MIRU-VNTR typing data for all 24 loci and categorical distance measure must be chosen for distance calculation. A phylogenetic lineage identification is obtained for 39 of the 97 strains when a similarity search is carried out by using the menu command ‘Automatically Set Best Matching Species and Lineage’ with a distance threshold of 0.17 (default value). By means of the tree-based identification, lineages of additional user strains can be derived. By drawing a NJ tree that is re-rooted at the branch containing the two M. canetti reference database strains (outgroup), species and lineage information can be reliably determined for 17 additional strains (one strain with lineage West African 1, three strains with lineage S, one strain with Beijing lineage, eight strains with lineage LAM and four strains with lineage EAI). A closer look at the remaining unidentified strains clearly reveals the presence of the two lineages (strains with ID 19, 25, 35, 40, 52, 42, 63 and ID 33, 78, 83, 56, 34, 72, 50, 51, 12, 79), which had been described as ‘Sierra Leone-1’ and ‘Sierra Leone-2’ in the original publication. The creation of a MST using the 24 MIRU-VNTR loci and a maximum locus difference of four within a CC reproduces the CCs of the publication, however, with a slightly different visualization and ordering. Remarkably, MST groups the ‘Sierra Leone-1’ and ‘Sierra Leone-2’ strains into specific CCs. The key findings of Homolka et al. (28) are thus confirmed by using MIRU-VNTRplus. The data exploration table gives an overview of all strains and allows further data exploration. Possible new MtbC15-9 types can be submitted by choosing the menu command ‘Assign MLVA MtbC15-9 Types’.
CONCLUSION
The MIRU-VNTRplus application allows quick identification of lineages for MTBC strains and exploration of data based on a combination of up to four different genotyping methods. Furthermore, an expanding nomenclature service for MLVA MtbC types has been established. It is planned to extend the reference database by adding further quality-controlled data sets. Future developments will include an open, extendable database with data sets from other researchers that can be used in addition to the reference database. Since MLVA, CRISPR and SNP typing schemes are being published for an increasing number of especially monomorphic pathogenic bacterial species, MIRU-VNTRplus may serve as a model for a powerful, generalized tool to analyse genotypes using these markers and other categorical data.
FUNDING
German Federal Ministry of Education and Research in the framework of the Network Zoonoses (grant number 01KI07124 to D.H. for development of the SOAP-interface); PathoGenomikPlus Network (grant number 0313801J to S.N. for collecting strains and genotyping). Funding for open access charge: German Federal Ministry of Education and Research (grant number 01KI07124 to D.H.).
Conflict of interest statement: P. Supply has declared a potential conflict of interest. P. Supply is a consultant for Genoscreen, Lille, France. All other authors have declared that no competing interests exist.
REFERENCES
- 1.World Health Organization. WHO Report. Geneva, Switzerland: World Health Organization; 2009. Global tuberculosis control. [Google Scholar]
- 2.Gagneux S, Small PM. Global phylogeography of Mycobacterium tuberculosis and implications for tuberculosis product development. Lancet Infect. Dis. 2007;7:328–337. doi: 10.1016/S1473-3099(07)70108-1. [DOI] [PubMed] [Google Scholar]
- 3.van Embden JD, Cave MD, Crawford JT, Dale JW, Eisenach KD, Gicquel B, Hermans P, Martin C, McAdam R, Shinnick TM. Strain identification of Mycobacterium tuberculosis by DNA fingerprinting: recommendations for a standardized methodology. J. Clin. Microbiol. 1993;31:406–409. doi: 10.1128/jcm.31.2.406-409.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sreevatsan S, Pan X, Stockbauer KE, Connell ND, Kreiswirth BN, Whittam TS, Musser JM. Restricted structural gene polymorphism in the Mycobacterium tuberculosis complex indicates evolutionarily recent global dissemination. Proc. Natl Acad. Sci. USA. 1997;94:9869–9874. doi: 10.1073/pnas.94.18.9869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hershberg R, Lipatov M, Small PM, Sheffer H, Niemann S, Homolka S, Roach JC, Kremer K, Petrov DA, Feldman MW, et al. High functional diversity in Mycobacterium tuberculosis driven by genetic drift and human demography. PLoS Biol. 2008;6:e311. doi: 10.1371/journal.pbio.0060311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brosch R, Gordon SV, Marmiesse M, Brodin P, Buchrieser C, Eiglmeier K, Garnier T, Gutierrez C, Hewinson G, Kremer K, et al. A new evolutionary scenario for the Mycobacterium tuberculosis complex. Proc. Natl Acad. Sci. USA. 2002;99:3684–3689. doi: 10.1073/pnas.052548299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gagneux S, DeRiemer K, Van T, Kato-Maeda M, de Jong BC, Narayanan S, Nicol M, Niemann S, Kremer K, Gutierrez MC, et al. Variable host-pathogen compatibility in Mycobacterium tuberculosis. Proc. Natl Acad. Sci. USA. 2006;103:2869–2873. doi: 10.1073/pnas.0511240103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hirsh AE, Tsolaki AG, DeRiemer K, Feldman MW, Small PM. Stable association between strains of Mycobacterium tuberculosis and their human host populations. Proc. Natl Acad. Sci. USA. 2004;101:4871–4876. doi: 10.1073/pnas.0305627101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Supply P, Magdalena J, Himpens S, Locht C. Identification of novel intergenic repetitive units in a mycobacterial two-component system operon. Mol. Microbiol. 1997;26:991–1003. doi: 10.1046/j.1365-2958.1997.6361999.x. [DOI] [PubMed] [Google Scholar]
- 10.Supply P, Allix C, Lesjean S, Cardoso-Oelemann M, Rüsch-Gerdes S, Willery E, Savine E, de Haas P, van Deutekom H, Roring S, et al. Proposal for standardization of optimized mycobacterial interspersed repetitive unit-variable-number tandem repeat typing of Mycobacterium tuberculosis. J. Clin. Microbiol. 2006;44:4498–4510. doi: 10.1128/JCM.01392-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kamerbeek J, Schouls L, Kolk A, van Agterveld M, van Soolingen D, Kuijper S, Bunschoten A, Molhuizen H, Shaw R, Goyal M, et al. Simultaneous detection and strain differentiation of Mycobacterium tuberculosis for diagnosis and epidemiology. J. Clin. Microbiol. 1997;35:907–914. doi: 10.1128/jcm.35.4.907-914.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Oelemann MC, Diel R, Vatin V, Haas W, Rüsch-Gerdes S, Locht C, Niemann S, Supply P. Assessment of an optimized mycobacterial interspersed repetitive- unit-variable-number tandem-repeat typing system combined with spoligotyping for population-based molecular epidemiology studies of tuberculosis. J. Clin. Microbiol. 2007;45:691–697. doi: 10.1128/JCM.01393-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wirth T, Hildebrand F, Allix-Béguec C, Wölbeling F, Kubica T, Kremer K, van Soolingen D, Rüsch-Gerdes S, Locht C, Brisse S, et al. Origin, spread and demography of the Mycobacterium tuberculosis complex. PLoS Pathog. 2008;4:e1000160. doi: 10.1371/journal.ppat.1000160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Comas I, Homolka S, Niemann S, Gagneux S. Genotyping of genetically monomorphic bacteria: DNA sequencing in Mycobacterium tuberculosis highlights the limitations of current methodologies. PLoS ONE. 2009;4:e7815. doi: 10.1371/journal.pone.0007815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Le Flèche P, Fabre M, Denoeud F, Koeck J, Vergnaud G. High resolution, on-line identification of strains from the Mycobacterium tuberculosis complex based on tandem repeat typing. BMC Microbiol. 2002;2:37. doi: 10.1186/1471-2180-2-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grissa I, Bouchon P, Pourcel C, Vergnaud G. On-line resources for bacterial micro-evolution studies using MLVA or CRISPR typing. Biochimie. 2008;90:660–668. doi: 10.1016/j.biochi.2007.07.014. [DOI] [PubMed] [Google Scholar]
- 17.Tang C, Reyes JF, Luciani F, Francis AR, Tanaka MM. spolTools: online utilities for analyzing spoligotypes of the Mycobacterium tuberculosis complex. Bioinformatics. 2008;24:2414–2415. doi: 10.1093/bioinformatics/btn434. [DOI] [PubMed] [Google Scholar]
- 18.Brudey K, Driscoll JR, Rigouts L, Prodinger WM, Gori A, Al-Hajoj SA, Allix C, Aristimuño L, Arora J, Baumanis V, et al. Mycobacterium tuberculosis complex genetic diversity: mining the fourth international spoligotyping database (SpolDB4) for classification, population genetics and epidemiology. BMC Microbiol. 2006;6:23. doi: 10.1186/1471-2180-6-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Allix-Béguec C, Harmsen D, Weniger T, Supply P, Niemann S. Evaluation and strategy for use of MIRU-VNTRplus, a multifunctional database for online analysis of genotyping data and phylogenetic identification of Mycobacterium tuberculosis complex isolates. J. Clin. Microbiol. 2008;46:2692–2699. doi: 10.1128/JCM.00540-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Niemann S, Harmsen D, Rüsch-Gerdes S, Richter E. Differentiation of clinical Mycobacterium tuberculosis complex isolates by gyrB DNA sequence polymorphism analysis. J. Clin. Microbiol. 2000;38:3231–3234. doi: 10.1128/jcm.38.9.3231-3234.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cavalli-Sforza LL, Edwards AW. Phylogenetic analysis. Models and estimation procedures. Am. J. Hum. Genet. 1967;19:233–257. [PMC free article] [PubMed] [Google Scholar]
- 22.Goldstein DB, Ruiz Linares A, Cavalli-Sforza LL, Feldman MW. Genetic absolute dating based on microsatellites and the origin of modern humans. Proc. Natl Acad. Sci. USA. 1995;92:6723–6727. doi: 10.1073/pnas.92.15.6723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shriver MD, Jin L, Boerwinkle E, Deka R, Ferrell RE, Chakraborty R. A novel measure of genetic distance for highly polymorphic tandem repeat loci. Mol. Biol. Evol. 1995;12:914–920. doi: 10.1093/oxfordjournals.molbev.a040268. [DOI] [PubMed] [Google Scholar]
- 24.Takezaki N, Nei M. Empirical tests of the reliability of phylogenetic trees constructed with microsatellite DNA. Genetics. 2008;178:385–392. doi: 10.1534/genetics.107.081505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sokal RR, Michener CD. A statistical method for evaluating systematic relationships. Univ. Kansas Sci. Bull. 1958;38:1409–1438. [Google Scholar]
- 26.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 27.Kruskal JB. On the shortest spanning subtree of a graph and the traveling salesman problem. Proc. Am. Math. Soc. 1956;7:48–50. [Google Scholar]
- 28.Homolka S, Post E, Oberhauser B, George AG, Westman L, Dafae F, Rüsch-Gerdes S, Niemann S. High genetic diversity among Mycobacterium tuberculosis complex strains from Sierra Leone. BMC Microbiol. 2008;8:103. doi: 10.1186/1471-2180-8-103. [DOI] [PMC free article] [PubMed] [Google Scholar]