Abstract
ToppCluster is a web server application that leverages a powerful enrichment analysis and underlying data environment for comparative analyses of multiple gene lists. It generates heatmaps or connectivity networks that reveal functional features shared or specific to multiple gene lists. ToppCluster uses hypergeometric tests to obtain list-specific feature enrichment P-values for currently 17 categories of annotations of human-ortholog genes, and provides user-selectable cutoffs and multiple testing correction methods to control false discovery. Each nameable gene list represents a column input to a resulting matrix whose rows are overrepresented features, and individual cells per-list P-values and corresponding genes per feature. ToppCluster provides users with choices of tabular outputs, hierarchical clustering and heatmap generation, or the ability to interactively select features from the functional enrichment matrix to be transformed into XGMML or GEXF network format documents for use in Cytoscape or Gephi applications, respectively. Here, as example, we demonstrate the ability of ToppCluster to enable identification of list-specific phenotypic and regulatory element features (both cis-elements and 3′UTR microRNA binding sites) among tissue-specific gene lists. ToppCluster’s functionalities enable the identification of specialized biological functions and regulatory networks and systems biology-based dissection of biological states. ToppCluster can be accessed freely at http://toppcluster.cchmc.org.
INTRODUCTION
One of the primary issues in analyzing large-scale biological data, like gene expression data, is the interpretation vis-à-vis the functional implications of the identified gene clusters. The availability of diverse functional annotations and molecular features associated with individual genes and shared across different gene groups often aids in the identification of critical biological properties associated with a biological state, process or response and can provide useful biological insight. Typical annotations include the Gene Ontology, biochemical pathways, protein–protein interactions, protein domain information and, in some cases, gene-disease associations. A wide variety of tools exist for overrepresentation analysis of functional annotations in single gene lists such as DAVID (1), FatiGO (2), g:Profiler (3) etc. A detailed review of the numerous functional analysis tools and the methods used is available in ref. 4, and recently in ref. 5. Recent approaches to such analyses, especially those that involve microarray-based gene expression data, employ the gene-set enrichment methods. Since the introduction of Gene Set Enrichment Analysis (6), such techniques have been highly successful in functional feature dissection of individual gene lists, for example in a gene expression data set among variably or similarly regulated gene lists. However, as we gain in our ability to detect complex correlated biological phenomena against the backdrop of ever-increasing knowledge of molecular and biological entities and properties, a critical goal must be that we also enhance our ability to identify components, activities and interactions responsible for systems functions and regulatory mechanisms such as has been pioneered in applications such as WGCNA (7) and ARACNE (8). Similarly, effective and efficient visualizations of data against current knowledge are critical to catalyze new hypotheses.
Here, we present an intuitive and efficient tool to analyze and visualize shared and specific features associated with any number of gene sets. We provide a workflow and tool suite that enables the co-analysis of the multiple gene lists in such a way as to preserve the relationships between gene lists and to provide secondarily analyzable data documents and visualizations capable of representing clusters of gene functions and features that are specific or shared among the multiple gene lists. For example, a multiple gene-list analysis approach could allow for the comparison of functional overrepresentations that differ as a function of time in time-series gene expression experiments, or in gene lists that reflect the relative differentiation of various tissue types or cell types. Currently, there are no existing tools that possess the capability to investigate multiple gene lists together and provide a functional modular map that includes a rich set of annotations. Progress in this area is shown in recent tools such as High-Throughput GoMiner (9) and GOEAST (10); however, these tools currently enable only Gene Ontology-based set enrichments and would be insensitive to regulatory mechanisms, protein–protein interactions, phenotypes, diseases, small molecule and other types of relationships that could enable useful inference. PageMan (11) is an innovative application that allows analysis of multiple microarray expression profile clusters at a time, with an intuitive heatmap visualization based on set enrichments to organism-specific user-generated ontology mapping files.
Most gene-set enrichment tools use the hypergeometric distribution as the statistical model to obtain the probability of a functional term occurring multiple times in a gene list just by chance. Here, ToppCluster makes use of the same method by means of the gene set enrichment functionality established in ToppGene (12) to assess significant feature enrichments in multiple input gene lists. Next, we utilize heatmaps or networks, both well-established visualization tools in genomics, to generate functional maps of the gene clusters. Heatmaps are highly suitable to display large data sets by means of color intensity values (13). By representing the significance of functional terms as the intensity of colors on the heatmap, we illustrate an extremely simple, yet effective way to visualize the biological functional theme of several gene sets at once. Additionally, we provide the feature of exporting the results for further analysis in various formats, including TreeView cluster trees (13,14), Cytoscape (15) and Gephi (16) compatible networks. ToppCluster is available as an open and freely accessible web application server at http://toppcluster.cchmc.org/. To demonstrate its usefulness, we used gene lists from the Tissue-specific Gene Expression and Regulation (TiGER) database (17) as inputs to ToppCluster to generate functional maps of features that are tissue-specific or shared between tissues. We show that ToppCluster allows for the identification of organ-specific phenotype associations, biological processes and genes whose mRNAs are targets of microRNAs (miRs) as well as genes whose promoters contain cis-elements for known transcription factors. Our goal is to provide a basis for researchers to assemble diverse knowledge and feature property connections across large data sets in such a way as to gain a deeper ability to model pathways and mechanisms responsible for systems function and to illuminate the functional relevance of their data in relation to high-dimensional human gene-associated knowledge.
METHODS
System workflow
The primary object of ToppCluster is the identification of biological themes in data sets involving numerous gene sets. A typical example is a time-series microarray experiment. The principal strength of ToppCluster lies in the ability to co-analyze multiple gene lists and to depict the results in a form that facilitates comparative and contrastive analysis.
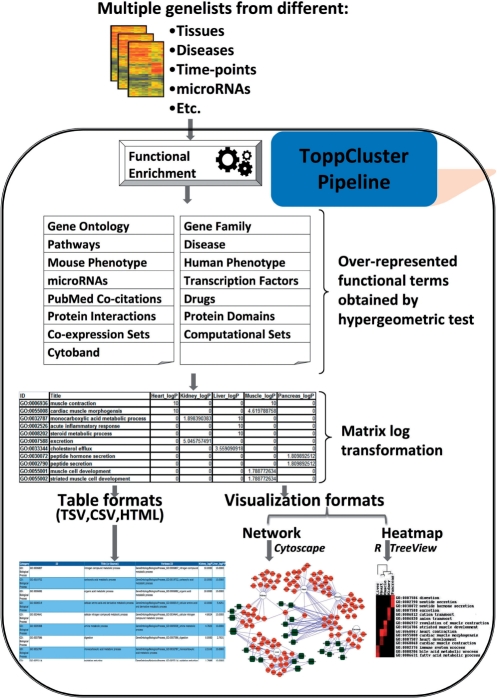
Figure 1 shows a schematic representation of the ToppCluster pipeline. The input consists of multiple gene lists from various experiments involving, for example, different tissues, time-points, cell-types, microRNA targets etc. P-value cutoff and the correction method chosen [Bonferroni, false discovery rate (FDR) or None] are used as filters. The user can select one or more annotation types to be included in the output. The enrichment functionality of the ToppGene suite (12) is used by ToppCluster to derive over-represented annotations. ToppGene contains 17 human gene-based annotation types, including Gene Ontology-Biological Process, Molecular Function, Cellular Component, Mouse Phenotype, Human Phenotype, Pathways, Transcription Factor Binding Sites, predicted MicroRNA targets, PubMed co-citations, Protein domains, Protein–Protein Interactions, Cytoband, Gene Coexpression, Expression Correlation (‘Computational’), Drug/Chemical and Disease. Links to data sources for these annotations in ToppGene can be found in the ‘Links’ section of the ToppCluster website. Additional details about the types and numbers of annotations can be found in the ‘Database Info’ section on the ToppCluster homepage under the ‘ToppGene’ header. After finalizing the input parameters, gene-associated feature enrichments are computed in ToppGene (12) based on the hypergeometric distribution test. The initial output is a result matrix that has columns that relate to each input gene list, and rows that represent the overrepresented features of any of the gene lists. One column for each named gene list is a significance value equal to the negative log of the P-value, and the other column for each gene list is a comma-delimited list of genes that have that feature (see below). If a given feature has a significant association for multiple gene list inputs, it is possible that there is an identical significance score, but completely different lists of genes that relate to that feature. The resulting functional enrichment matrix can be hierarchically clustered for visualization and analysis as a heatmap or transformed into a Cytoscape-compatible XGMML network format or a Gephi compatible GEXF network format. If the heatmap generation option is chosen, the functional enrichment matrix is subjected to two-dimensional hierarchical clustering (see below), where first the rows and then the columns are reordered according to similar scores. In the tabular format, the genes from the particular gene list contributing to the significance score are provided in an adjoining table. Third-party software can be used to import and visualize the heatmaps or networks. The networks can also be obtained as static images.
Figure 1.
Schematic representation of the ToppCluster pipeline. Multiple genelists are named by the user and submitted through the ToppCluster interface. 17 different categories of annotations are available as of April 2010. The user can choose the categories to be included, the P-value cutoff, a method of correction for false discovery and the type of output. Functional enrichment analysis is done on each gene list and the results are collated into a single matrix. Significance P-values are log transformed (−log10) into scores and the genes per list that intersect each enriched feature are placed into a separate column. The results are delivered in the chosen output format.
Data input and interface
ToppCluster accepts input in one of two ways: (i) as separate lists of genes which can be successively added and named, or (ii), using the ‘alternative entry’ method, as a two-column list with genes in the first column and the name of the gene list in the second column. Accepted input is limited to human genes at present. One or any of the 17 annotation sources can be used for feature enrichment analyses. Each feature analysis can be adjusted based on the P-value cutoff, the multiple testing correction method or the minimum and maximum number of genes present for each annotation type. For example, limiting enrichments to ontologies that have fewer associated genes can allow for a greater focus on specific classes of gene feature or function. Multiple choices are available for the formatting and delivery of results. The user can opt for results to be obtained in tabular format as comma-separated values, tab-separated values or HTML table format. It is also possible to obtain the results in various visualization formats—a standard heatmap in a PDF file generated using R (18) (http://www.R-project.org), TreeView (13,14) clustered data tree (CDT) heatmap files, GenePattern (19) GCT format, Cytoscape (15) XGMML importable network formats, Gephi (16) importable GEXF network formats or as pre-laid out network images using the PNG option.
Generation of enrichment data map
Each labeled gene list is fed to the ToppGene (12) web service. The functional enrichment results for each gene list for the selected categories are then compiled and concatenated into a tabular format. Here, we have used a new approach to represent the significance of the functional term in a gene list. We take the negative logarithm (−log10) of the P-value corresponding to the term, thus obtaining ‘significance scores’ above zero. We round off any values over 10 to 10. Hence, the functional terms corresponding to a gene list have significance scores falling between 0 and 10. If the heatmap option is chosen, the matrix is subjected to two-dimensional hierarchical clustering using the Euclidean distance measure with average linkage via R (18).
Visualization
ToppCluster results can be viewed graphically as heatmaps or networks. The ToppCluster heatmap-based output can be obtained in two ways: as a heatmap image in a PDF format file, or as a set of files compatible with the TreeView (13,14) software. TreeView (13,14) can be used to open the clustered data files to generate an interactive heatmap view. Heatmap images can subsequently be saved from TreeView (13,14).
ToppCluster results can also be exported as Cytoscape (15) supported network file types [XGMML; choose ‘Import’, “Network (multiple file types)”.], Gephi supported network files (GEXF) or as static images (PNG). An interactive HTML output format lets the user select features of interest from the results to be included in the network. Following this, the user is allowed to select the type, layout and file-format for the network. The network can be displayed in two very different ways: a ‘Gene Level’ option generates the entire network including the genes, while an ‘Abstracted’ option excludes the genes from the network, retaining only the enriched terms as nodes that are related to the input gene lists via edge relationships that subsume the list of specific genes. In this option, the network shows the input gene lists connected to the enriched terms by weighted edges; the edge-weight is set to the significance score of the enriched term, and the list of genes are available as an annotation field from Cytoscape’s data panel window in the Edge Attribute Browser.
Implementation
ToppCluster is a distributed system implemented in Java that runs across a cluster of Linux servers utilizing the Sun Glassfish Enterprise Server environment. ToppCluster passes data to ToppGene via Java Messaging Services (JMS). JMS automatically distributes all gene-list enrichment jobs to available ToppGene enrichment analysis nodes. The TreeView clustered data files and the PDF heatmap are generated using embedded R (18) scripts that run as scheduled jobs on the CCHMC Computational Cluster (http://bmi.cchmc.org/resources/clusters). Network images are generated using the JAVA JUNG (20) libraries for analysis and visualization of network data. ToppCluster uses the jQuery AJAX Library for dynamic HTML-based user interfaces.
UTILITY OF TOPPCLUSTER
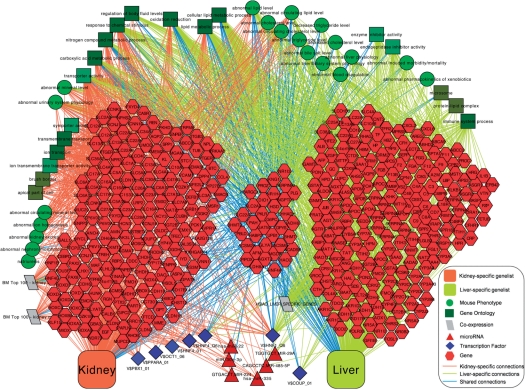
We demonstrate the efficacy of ToppCluster using a simple example—a set of tissue specific gene lists. Tissue-specific gene lists from the TiGER (17) database were selected for genes most highly expressed in heart, muscle, liver, kidney and pancreas. From these, we sought to identify and partition out the tissue-specific gene lists based on their shared and specific disease phenotypes and potential regulatory mechanism relationships. The formatted and labeled lists were then submitted to ToppCluster with a P-value cutoff of 0.05 and FDR correction method. The features selected were Mouse Phenotype, microRNA and Transcription Factor Binding Sites. Importantly, no false discovery correction method was applied to microRNAs as the hypothesis for its role is not based on a genome-wide relative enrichment, but rather a Boolean true–false question as to whether the miRNA is expressed and what genes it might target. First, the ‘Abstracted’ network option was used to generate a Cytoscape-compatible network file containing all enriched term relationships. Figure 2A shows the network exported as an image from Cytoscape after using the Spring Embedded Layout function and the significance-based edge weights. Some of the shared and specific phenotypes, microRNAs,and over-represented transcription factors are labeled in the figure.
Figure 2.
(A) An ‘Abstracted’ Network showing enriched Mouse Phenotype terms, microRNAs and transcription factors associated with five tissue-specific gene clusters—heart, kidney, liver, muscle and pancreas. (B) Dissected gene-level view of enriched Mouse Phenotype terms, microRNAs and Transcription Factors shared between the kidney and liver specific gene lists. MP, Mouse Phenotype; TFBS, transcription factor binding site; miRNA, microRNA.
From the Abstracted network view, there are distinct functional separations. Notably, the liver gene list shows highly significant enrichments for sets of genes that confer specific phenotypes such as abnormal liver/biliary morphology, decreased circulating cholesterol and abnormal blood coagulation. The heart and skeletal muscle lists share general cardiac muscle contractility and morphology phenotypes, but differ with respect to phenotypes that include abnormal impulse conducting system, irregular heartbeat and dilated atria for the heart, and decreased muscle mass, abnormal muscle development and muscle weakness for skeletal muscle. Also shared among the two are the heart- and muscle-expressed transcription factors myocyte enhancer factor-2 (MEF-2) (21) and serum response factor (22), but miR-29a,b,c and miR-100 –targeted genes are significantly enriched. Consistent with this, miR-29 has been shown to be a critical suppressor of cardiac fibrosis (23). The kidney genes show extensive relationship enrichments to abnormalities of kidney structure and function, as well as transporter-associated specific functions such as excretion and ion transport. The transcription factor Pou3f3, the absence of which causes multiple kidney phenotypes, binds to OCT class transcription factor binding sites, which are over-represented in the kidney genes. The kidney gene list is also enriched for promoter transcription factor binding sites for PBX1, which also regulates nephrogenesis and uretric branching (24). Consistent with existing knowledge, hepatocyte nuclear factors HNF1 and HNF4 are shared between the liver and kidney genes (25,26). The liver-specific genes are enriched for the Chicken Ovalbumin Upstream Promoter-Transcription Factor (COUP-TF). This transcription factor, although usually expressed ubiquitously especially during development, is found to be highly similar in its binding site to the liver-expressed Hepatocyte Nuclear Factor-4 (HNF-4). The expression of COUP-TF in almost all tissues and the fact that that it acts as a repressor for the liver-specific gene Orinthine Transcarbamylase (OTC) led to the theory that it may act as a repressor of liver specific genes in other tissues (27). The pancreatic genes show phenotypes such as disorganized pancreatic islets, abnormal pancreas development and abnormal insulin secretion. The kidney genes share circulating amino acid, cholesterol, lipid and mineral level phenotypes with the liver genes. The pancreas-specific genes show enrichment for the transcription factor GATA1, which is known to be involved in cell-specific regulation of genes in multiple endocrine organs including the pancreas (28). Also of interest is the microRNA miR-190, which is specific to the pancreas genes. miR-190 has been found to be significantly upregulated in pancreatic cancer tissues and cell lines (29).
To provide a dissected gene-level view of some of these terms, we chose the phenotypes and transcription factors shared between the liver and kidney genes. Using the ‘Gene Level’ network option, we generated a Cytoscape compatible network showing only the genes, phenotypes and transcription factors shared between the two sets, as shown in Figure 2B. ToppCluster allows the user to select terms of interest to be included in the network. This would be especially useful when an investigator wants to further explore only some of the enriched terms in the output. This feature was used to generate Figure 3, where terms enriched in multiple categories were selected and the gene level network was generated.
Figure 3.
Gene-level network showing user-selected enriched terms from Gene Ontology, Mouse Phenotype, Co-expression, microRNAs and transcription factors for the kidney and liver-specific gene lists.
The above use case, although recapitulating existing knowledge, demonstrates the ability of ToppCluster to tease out the shared and specific functions and regulatory elements among multiple gene clusters.
An Excel file containing the input gene lists, the Cytoscape network session (CYS) file and the network data text file are available in the ‘Supplementary’ section of the ToppCluster homepage. Also available is a heatmap view corresponding to Figure 2A generated using TreeView and the TreeView format clustered data files.
LIMITATIONS
As mentioned in the review (5), one of the problems with cross-comparing enrichment analyses from multiple gene lists is that the sizes of the gene lists affect the FDR-corrected enrichment P-values. This can make it somewhat difficult to compare the P-values across gene lists when their sizes are considerably different. An algorithm to offset these differences that makes these comparisons more accurately may be called for. In addition, the specific hypotheses and relationship assumptions that underlie the use of false discovery P-value correction in the significance testing of some categorical feature enrichments in a given gene list is a complex subject. Some circumstances may warrant a consideration of analysis results obtained without the use of false discovery P-value correction. This is certainly the case for considering the potential involvement of miRs in regulatory networks, where the primary consideration may be factoring of knowledge of whether any of the miRs that could target critical genes are actually expressed. A key consideration beyond the scope of this manuscript is therefore what are the causal or relationship models that are being sought out, and when is it appropriate to provide sensitivity versus specificity with respect to the detection of relevant relationships among a set of genes that determine function in a complex system? These considerations may considerably affect the optimal choice of false discovery method and appropriate statistical cutoff. In addition, lists formed from biased gene analysis methods are not corrected in ToppGene because our reference sets are not correctable at this time (4). However, differential significance results between lists should be relatively resistant to this effect for lists that contain similar numbers of genes.
CONCLUSION
Existing gene set enrichment analysis tools cover a wide range of annotations and aid in the analysis of gene lists of interest identified from large-scale experiments (30). However, the novel concept presented in this paper and achieved by the ToppCluster web server is that to tease out highly significant relationships among a set of co-acting or cooperating genes in one context, it can be extremely valuable to compare these with those of genes from other biological states. ToppCluster does this by comparative co-analysis of multiple gene sets. In this presentation, we have demonstrated the utility and potential for discovery offered by ToppCluster to identify biological processes and putative regulatory mechanisms associated with human tissue-specific gene expression gene sets that exhibit rich disease and phenotypes impacts. We envision ToppCluster-enabled workflows of analysis, prediction and discovery as providing a valuable tool for researchers seeking to dissect complex biological processes to hone in on specific genes, pathways and regulatory mechanisms for prediction and further experimentation of systems function, dysfunction and therapeutic agent effects. The ability to visualize functional relationships across multiple gene lists provides, in our opinion, novel opportunities to form new hypotheses about the roles and interactions of underlying biological mechanisms responsible for the determination of biological states including development, homeostasis and disease pathology.
FUNDING
National Institutes of Health/National Institute of Diabetes and Digestive and Kidney Diseases (NIH/NIDDK) 1U01 DK70219 (Murine Atlas of a Genitourinary Development Molecular Anatomy Project); PHS Grant P30 DK078392 (Cincinnati Digestive Health Center); U54 RR025216 (CTSA: Cincinnati Center for Clinical and Translational Sciences); and U01DE020049 NIDCR (FACEBASE Consortium). Funding for open access charge: National Institutes of Health grants.
Conflict of interest statement. None declared.
REFERENCES
- 1.Dennis G., Jr, Sherman B.T., Hosack D.A., Yang J., Gao W., Lane H.C., Lempicki R.A. DAVID: Database for annotation, visualization, and integrated discovery. Genome Biol. 2003;4:P3. [PubMed] [Google Scholar]
- 2.Al-Shahrour F., Diaz-Uriarte R., Dopazo J. FatiGO: a web tool for finding significant associations of Gene Ontology terms with groups of genes. Bioinformatics. 2004;20:578–580. doi: 10.1093/bioinformatics/btg455. [DOI] [PubMed] [Google Scholar]
- 3.Reimand J., Kull M., Peterson H., Hansen J., Vilo J. g:Profiler–a web-based toolset for functional profiling of gene lists from large-scale experiments. Nucleic Acids Res. 2007;35:W193–W200. doi: 10.1093/nar/gkm226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Khatri P., Draghici S. Ontological analysis of gene expression data: current tools, limitations, and open problems. Bioinformatics. 2005;21:3587–3595. doi: 10.1093/bioinformatics/bti565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Huang da W., Sherman B.T., Lempicki R.A. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S., et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl Acad. Sci. USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Langfelder P., Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinform. 2008;9:559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Margolin A.A., Nemenman I., Basso K., Wiggins C., Stolovitzky G., Dalla Favera R., Califano A. ARACNE: an algorithm for the reconstruction of gene regulatory networks in a mammalian cellular context. BMC Bioinform. 2006;7(Suppl. 1):S7. doi: 10.1186/1471-2105-7-S1-S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zeeberg B.R., Qin H., Narasimhan S., Sunshine M., Cao H., Kane D.W., Reimers M., Stephens R.M., Bryant D., Burt S.K., et al. High-Throughput GoMiner, an ‘industrial-strength’ integrative gene ontology tool for interpretation of multiple-microarray experiments, with application to studies of Common Variable Immune Deficiency (CVID) BMC Bioinform. 2005;6:168. doi: 10.1186/1471-2105-6-168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zheng Q., Wang X.J. GOEAST: a web-based software toolkit for Gene Ontology enrichment analysis. Nucleic Acids Res. 2008;36:W358–W363. doi: 10.1093/nar/gkn276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Usadel B., Nagel A., Steinhauser D., Gibon Y., Blasing O.E., Redestig H., Sreenivasulu N., Krall L., Hannah M.A., Poree F., et al. PageMan: an interactive ontology tool to generate, display, and annotate overview graphs for profiling experiments. BMC Bioinform. 2006;7:535. doi: 10.1186/1471-2105-7-535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen J., Xu H., Aronow B.J., Jegga A.G. Improved human disease candidate gene prioritization using mouse phenotype. BMC Bioinform. 2007;8:392. doi: 10.1186/1471-2105-8-392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eisen M.B., Spellman P.T., Brown P.O., Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc. Natl Acad. Sci. USA. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Saldanha A.J. Java Treeview–extensible visualization of microarray data. Bioinformatics. 2004;20:3246–3248. doi: 10.1093/bioinformatics/bth349. [DOI] [PubMed] [Google Scholar]
- 15.Shannon P., Markiel A., Ozier O., Baliga N.S., Wang J.T., Ramage D., Amin N., Schwikowski B., Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bastian M., Heymann S., Jacomy M. Gephi: An Open Source Software for Exploring and Manipulating Networks; International AAAI Conference on Weblogs and Social Media; California: San Jose; 2009. [Google Scholar]
- 17.Liu X., Yu X., Zack D.J., Zhu H., Qian J. TiGER: a database for tissue-specific gene expression and regulation. BMC Bioinform. 2008;9:271. doi: 10.1186/1471-2105-9-271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.R-Development-Core-Team. R Foundation for Statistical Computing. Vienna: Austria; 2007. R: a language and environment for statistical computing. [Google Scholar]
- 19.Reich M., Liefeld T., Gould J., Lerner J., Tamayo P., Mesirov J.P. GenePattern 2.0. Nat. Genet. 2006;38:500–501. doi: 10.1038/ng0506-500. [DOI] [PubMed] [Google Scholar]
- 20.Madadhain J., Fisher D., Smyth P., White S., Boey Y. Analysis and visualization of network data using JUNG. J. Stat. Soft. 2005;10:1–35. [Google Scholar]
- 21.Naya F.J., Olson E. MEF2: a transcriptional target for signaling pathways controlling skeletal muscle growth and differentiation. Curr. Opin. Cell. Biol. 1999;11:683–688. doi: 10.1016/s0955-0674(99)00036-8. [DOI] [PubMed] [Google Scholar]
- 22.Miano J.M. Serum response factor: toggling between disparate programs of gene expression. J. Mol. Cell. Cardiol. 2003;35:577–593. doi: 10.1016/s0022-2828(03)00110-x. [DOI] [PubMed] [Google Scholar]
- 23.van Rooij E., Sutherland L.B., Thatcher J.E., DiMaio J.M., Naseem R.H., Marshall W.S., Hill J.A., Olson E.N. Dysregulation of microRNAs after myocardial infarction reveals a role of miR-29 in cardiac fibrosis. Proc. Natl Acad. Sci. USA. 2008;105:13027–13032. doi: 10.1073/pnas.0805038105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schnabel C.A., Godin R.E., Cleary M.L. Pbx1 regulates nephrogenesis and ureteric branching in the developing kidney. Dev. Biol. 2003;254:262–276. doi: 10.1016/s0012-1606(02)00038-6. [DOI] [PubMed] [Google Scholar]
- 25.Pontoglio M. Hepatocyte nuclear factor 1, a transcription factor at the crossroads of glucose homeostasis. J. Am. Soc. Nephrol. 2000;11(Suppl. 16):S140–S143. [PubMed] [Google Scholar]
- 26.Taraviras S., Monaghan A.P., Schutz G., Kelsey G. Characterization of the mouse HNF-4 gene and its expression during mouse embryogenesis. Mech. Dev. 1994;48:67–79. doi: 10.1016/0925-4773(94)90017-5. [DOI] [PubMed] [Google Scholar]
- 27.Kimura A., Nishiyori A., Murakami T., Tsukamoto T., Hata S., Osumi T., Okamura R., Mori M., Takiguchi M. Chicken ovalbumin upstream promoter-transcription factor (COUP-TF) represses transcription from the promoter of the gene for ornithine transcarbamylase in a manner antagonistic to hepatocyte nuclear factor-4 (HNF-4) J. Biol. Chem. 1993;268:11125–11133. [PubMed] [Google Scholar]
- 28.Viger R.S., Guittot S.M., Anttonen M., Wilson D.B., Heikinheimo M. Role of the GATA family of transcription factors in endocrine development, function, and disease. Mol. Endocrinol. 2008;22:781–798. doi: 10.1210/me.2007-0513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang Y., Li M., Wang H., Fisher W.E., Lin P.H., Yao Q., Chen C. Profiling of 95 microRNAs in pancreatic cancer cell lines and surgical specimens by real-time PCR analysis. World J. Surg. 2009;33:698–709. doi: 10.1007/s00268-008-9833-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kann M.G. Advances in translational bioinformatics: computational approaches for the hunting of disease genes. Brief. Bioinform. 11:96–110. doi: 10.1093/bib/bbp048. [DOI] [PMC free article] [PubMed] [Google Scholar]