Abstract
Early innate events that enable priming of anti-fungal CD4 T cells are poorly understood. We engineered an attenuated fungal vaccine with a model epitope EαRFP to track vaccine immunity to Blastomyces dermatitidis comprehensively during yeast recognition, Ag presentation, and priming of naïve T cells. After subcutaneous injection of the vaccine, monocyte-derived inflammatory dendritic cells (DCs) are the earliest and largest population that associates with yeast, carrying them into the draining nodes. Despite marked association with yeast, these DCs fail to display surface peptide:MHC complexes or prime naïve T cells. Instead, the ability to display Ag and prime CD4 T cells resides with LN resident DCs after Ag transfer from immigrant DCs, and with skin migratory DCs. Our work reveals the dynamic interplay among distinct DC subsets that prime naïve CD4 T cells after yeast are injected in the skin, and discloses the cellular elements that induce vaccine immunity to fungi.
Keywords: TCR tg mouse, Vaccine Immunity, Dendritic Cells, T-cell priming, Fungi
INTRODUCTION
An increase in fungal diseases has fueled scientific interest in fungal pathogenesis and antifungal immunity, but a vaccine for any of the human systemic mycoses has never been developed. We genetically engineered an attenuated vaccine against experimental infection with the dimorphic fungus Blastomyces dermatitidis, the agent of blastomycosis. Inhalation of spores into the lung of a mammalian host promotes a phase transition to the pathogenic yeast form (Bradsher, 1988), which produces pneumonia or disseminated infection depending on the host’s immune status. In a murine model, intra-tracheal administration of spores or yeast leads to pneumonia with 100% mortality. Subcutaneous (s.c.) administration of the attenuated vaccineprotects mice against experimental infection and induces sterilizing immunity (Wüthrich et al., 2000).
The development of fungal vaccines requires an understanding of the elements that bridge innate and adaptive responses during the generation of anti-fungal immunity. CD4+ T cells mediate the control and clearance of fungi, with Th1 responses having a requisite role, including in vaccine immunity (Romani, 2008). During the earliest stages of fungal infection, pattern recognition receptors expressed on cells of the myeloid lineage enable the host to first sense the presence of pathogenic fungi and fungal surfaces (Brown, 2006; Netea et al., 2008). Myeloid cells, particularly dendritic cells (DCs), bridge innate and adaptive immune response to pathogens across all kingdoms. Because of this, DCs have become a target for novel vaccine development strategies (Steinman, 2008).
Dendritic cells are a heterogeneous population of Ag-presenting cells (APCs) that drive immune responses toward inflammatory or regulatory pathways (Banchereau and Steinman, 1998). Adaptive immunity is regulated and fine-tuned by DCs, depending on the type of Ag and DC subset residing at the site of pathogen entry (Dudziak et al., 2007). In the skin draining lymph nodes (LNs), which represent the chief site of T cell priming after s.c. vaccination, there are two major categories of DCs: migratory DCs and LN-resident DCs (Villadangos and Schnorrer, 2007). Migratory DCs include skin-resident DCs and monocyte-derived inflammatory DCs. Skin DCs are comprised of subsets of dermal DCs (CD103+ and CD103−) and Langerhans cells (Kaplan et al., 2008), whereas monocyte-derived DCs arise when circulating monocytes differentiate into DCs at the site of inflammation (Leon et al., 2007).
The roles served by these distinct subsets of DCs in priming T cell responses have been analyzed in response to soluble model Ags, viruses, bacteria and parasites, but they are less well understood for the fungi. In Leishmania major cutaneous infection models, and in systems using model, soluble proteins such as EαRFP injected s.c., skin DCs are key players in controlling infection or inducing T cell responses (Itano et al., 2003; Moll et al., 1993). Skin DCs also regulate hypersensitivity responses (Kaplan et al., 2005) by taking up foreign or self-proteins at the site of infection, carrying the cargo to the draining LN and presenting the Ags to T cells (Heath et al., 2004). Skin migratory DCs also may cooperate with LN resident DCs to induce T cell responses by transferring viral Ag to resident DCs (Allan et al., 2006), and in response to OVA, allowing resident DCs to trap Ag specific T cells (Allenspach et al., 2008) while migratory DCs induce proliferation (Allan et al., 2006) Hence, LN-resident DCs also contribute to. presenting Ag in the skin draining LNs (Iezzi et al., 2006; Lee et al., 2009).
Monocyte-derived DCs have gained interest in priming T cell responses to microbes (Hohl et al., 2009; Serbina et al., 2008). Monocyte-derived inflammatory DCs were shown to prime Th1 responses in a model of cutaneous Leishmaniasis (Leon et al., 2007) and to prime Ag-specific CD4+ Tg cells in an experimental model of pulmonary Aspergillosis(Hohl et al., 2009). In Aspergillus infection, CCR2+ Ly6Chi monocyte-derived DCs were necessary for priming T-cells in the lung, but not in the spleen, underscoring the differences in how T-cells are primed in different tissue compartments. The studies above clarify how distinct DC subsets prime T cells in response to model or non-fungal agents, or to fungi that are inhaled into the lung during primary infection. They do not offer insight into the early cellular dynamics that underpin generation of a vaccine-induced immune response to fungi, especially initiated in the skin.
We engineered vaccine yeast to express the model Ag EαRFP (Rudensky et al., 1991) to assess sequential stages during the induction of vaccine immunity to fungi after s.c injection. We show that monocyte-derived inflammatory DCs initially take up the majority of vaccine yeast and traffic them to the draining LNs. A subsequent, smaller wave of vaccine-Ag trafficking to the LN is mediated by skin-derived DCs. Despite a large association of yeast with monocyte-derived DCs, the direct priming of naïve Ag-specific CD4 T cells in vivo is governed by LN-resident DCs and skin-derived DCs, whereas monocyte-derived DCs function mainly as carriers of Ag particles to the draining LN.
RESULTS
DC subsets that associate with vaccine yeast in the draining LNs
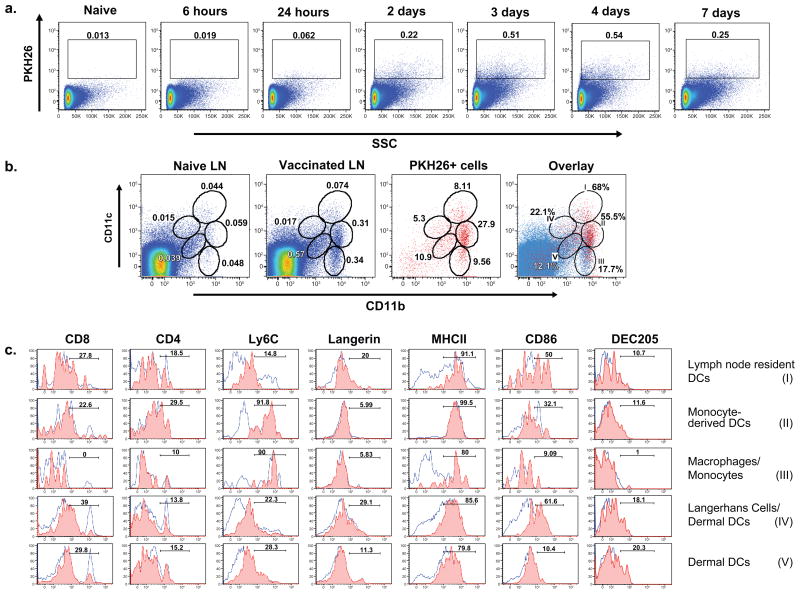
We tracked, analyzed and characterized the cells that associate with attenuated vaccine yeast in the skin-draining LNs by staining the yeast with the fluorescent cell linker PKH26. Thus, the cells that associate with vaccine yeast in the draining LNs are PKH26+. To elucidate the kinetics of yeast or yeast-Ag delivery into the draining LNs, mice were vaccinated s.c., and the draining LNs were harvested at serial time points. PKH26+ cells entered the LNs at 24 hours, and peaked two to three days later (Fig. 1a). Itano et al. (Itano et al., 2003) found that soluble protein injected s.c. entered the draining nodes 30 minutes later. This rapid entry was due to non-cell associated delivery of the Ag via the lymphatics. In contrast, yeast PKH26 signal in the node was delayed until 24 hours after vaccination, suggesting that yeast “particles” are actively transported by migratory cells rather than by passive movement of soluble fungal Ag into the draining node (Fig 1a).
Figure 1. Appearance of yeast in the skin-draining LNs of vaccinated mice.
Mice were vaccinated once s.c. with 107 PKH26+ yeast. (A) LNs were collected 6 hrs to 8 d after vaccination, and analyzed for cells containing PKH26+ yeast. Cells were gated on FSC/SSC, followed by a live-cell gate based upon violet fixable live/dead stain. (B) Five populations of APCs with PKH26+ yeast were identified based on CD11c/CD11b expression in LN cells 72 hrs after vaccination (3rd panel). The distribution of PKH26+ yeast among cells is indicated by the percent of PKH26+ cells in each gate. The fraction of LN cells in each gate is shown in vaccinated mice (2nd panel) and unvaccinated controls (1st panel). In the far right panel, PKH26+ cells (red) were superimposed on total LN cells (blue) to depict the frequency of Ag+ cells in each gate at the peak of influx 72 hrs after vaccination. (C) Identity of LN cells with PKH26+ yeast. Name and roman numeral in the right margin correspond to gates in panel B. The red and blue populations of LN cells were analyzed separately. The red histogram shows the phenotype of PKH26+ cells in the respective subset in panel B for vaccinated mice; the blue one shows the phenotype of the respective subset under non-inflammatory conditions (unvaccinated mice). Results in panels A, B and C are representative of 5 independent experiments.
Since DCs prime naïve T cells in the draining LNs, we sought to characterize the DCs that harbor and present yeast Ag(s) at this site. Multiple subsets of DCs have been defined by differences in the expression of surface markers (Lopez-Bravo and Ardavin, 2008; Villadangos and Schnorrer, 2007). We used CD11c/CD11b to identify 5 potential populations of DCs that contain yeast (Fig. 1b). By FACS analysis, we found a strong correlation between CD11b+ APC populations and PKH26. More than 90% of PKH26+ cells are CD11b+, and none are present in the CD11c single-positive population (Fig 1b). We created gates among the populations of PKH+ cells that correspond to established populations of DCs (Lopez-Bravo and Ardavin, 2008). We used additional markers (Fig. 1c) to confirm and further characterize the identities of the five populations of yeast-loaded cells near the peak of influx into the LN (~day 3). Population I corresponds to conventional, LN-resident DCs, which express high levels of CD11c and MHCII and include both CD8+ and CD8− subsets (Villadangos and Schnorrer, 2007). We used skin-FITC painting, which tracks migratory skin DC populations, to establish that population I is FITC− and confirm that this is the LN--resident DC population (Fig. S1a). In contrast, populations IV and V are migratory skin DCs. Skin DCs are a heterogeneous population of several subsets of dermal DCs and Langerhans cells, identifiable with DEC205 and Langerin markers (Segura and Villadangos, 2009). About 20% of cells in populations IV and V displayed these markers (Fig. 1c). These cells also were FITC+ after skin painting (Fig. S1a), supporting their identity as migratory skin DCs (Allan et al., 2006; Iezzi et al., 2006). We used two alternate means of tracking yeast associated with DCs to verify the findings with PKH26: 1) vaccine yeast engineered with red fluorescent protein and 2) the cell permeable dye CFSE. Each method yielded results qualitatively similar to those with PKH26, notably in the distribution of yeast among DC subsets (Fig. S1b). Due to the sensitivity and stability of PKH26, we used it for the rest of our experiments.
Monocyte-derived, inflammatory DCs are blood monocytes that differentiate into DCs upon inflammation. They express a monocyte Ly6C marker and exhibit a DC-like CD11cint/+ phenotype (Leon et al., 2007; Nakano et al., 2009). Populations II and III are Ly6C+ and >90% CD11bhi, indicating they are monocyte-derived cells (Fig. 1c). Population II also expresses CD11c, indicating they are likely monocyte-derived DCs. In contrast, population III expresses low or no CD11c and is thus likely to be infiltrating monocytes or macrophages. Overall, our phenotypic analysis showed that yeast associated with 3 major populations of DCs in the draining LNs of vaccinated mice: LN-resident DCs (population I), monocyte-derived DCs (population II) and skin-derived DCs (populations IV and V).
The kinetics and dynamics of vaccine yeast-associated cell populations in the draining LNs
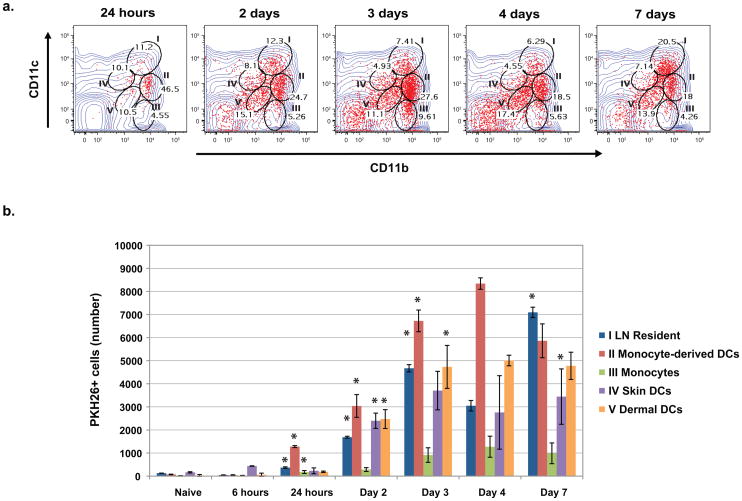
To investigate the kinetics and dynamics of DC subset trafficking of vaccine yeast into the draining LNs, we analyzed the distribution of PKH26+ cells serially over seven days. Monocyte-derived DCs are the first population to enter the LN with PKH26+ yeast. They appear as a dense population at 24 hours by FACS, when they account for 47% of the cells associated with vaccine yeast (Fig. 2a). These inflammatory DCs represent the single largest population of cells associated with yeast in the nodes, both in terms of the distribution of PKH26+ yeast among DCs and the numbers of cells with yeast in the node (Fig. 2a and b). Skin DC subsets (IV and V) with vaccine yeast appear in the node one day after monocyte-derived DCs (Fig. 2b), and account for ≈25% of cells associated with vaccine yeast by 48–72 hours after vaccination (Fig. 2a). During the first 72 hours after vaccination, PKH26+ yeast steadily associate with resident, conventional DCs (Fig 2b). Since this population (I) is not migratory, the shift in the PKH26+ signal to these resident DCs implies that migratory cells may transfer vaccine Ag to resident cells within the draining LN. Monocytes or macrophages (population III) increased slowly during vaccination, but accounted for only a small number of yeast-associated cells in the LN. A PKH26+ CD11c−CD11b− population also accumulated in the LN over time. This population is debris from dying PKH26+ cells because of its small size on the forward/side scatter analysis and lack of staining for cellular markers (data not shown). Neutrophils made up a small percentage of CD11b+ cells in the LN, but few vaccine yeast were associated with these cells (Fig. S2a). Thus, the chief cell populations that associated with vaccine yeast in the draining LNs are monocyte-derived, inflammatory DCs, skin-derived DCs and resident, conventional DCs. The temporal appearance of yeast with monocyte- and skin-derived DCs before they associate with resident DCs is consistent with the possibility that vaccine Ag is transferred to resident DCs in the LN. At the 72-hour peak of PKH26+ cell influx, the number of yeast-associated APCs is distributed evenly among three to four populations, suggesting that multiple DC subsets participate in the vaccine-induced protective immune response to fungi.
Figure 2. The dynamic distribution of yeast-Ag loaded cells in the draining LNs.
Mice were analyzed serially for 7 d after vaccination. LN cells withPKH26 + yeast (red) were analyzed and gated according to expression of CD11c and CD11b. (A) Gates and roman numerals correspond to those in Fig. 1. The changing distribution of PKH26+ yeast and percent of PKH26+ cells in each gate and population is depicted over time. (B) The distribution of PKH26+ yeast among the LN cell populations is shown in absolute numbers at each time point. Data are the mean ± SD of 2 independent experiments each with 4 mice per time point or group. *, p <0.05 denotes the subset increased significantly at that time point vs. the preceding one.
We considered that the distribution of live vaccine yeast among DC subsets could be affected by replicating yeast in the LN. We cultured the LNs of mice 72 hours after vaccination, but found <10 yeast/animal (limit of detection). We also compared the distribution of vaccine yeast among DCs in the LN after vaccination with live or heat-killed yeast and it was similar (Fig. S2b). Thus, the distribution likely reflects delivery of yeast into the LN, not replication in the node. We also evaluated whether the appearance of yeast Ag within LN-resident cells required ongoing delivery of yeast after vaccination. Pertussis toxin administered together with the vaccine to block cell migration halted entry of yeast into the LN (Fig. S2c). In contrast, when toxin was given 24 hours after vaccination, yeast still appeared in all of the DC subsets, including LN-resident DCs at 72 hours. These results suggest that cells from the vaccine site deliver yeast into the node, and may transfer them to LN-resident DCs.
Alterations of vaccine delivery into the LNs of CCR2−/−, CCR7−/−, and CD11cDTR mice
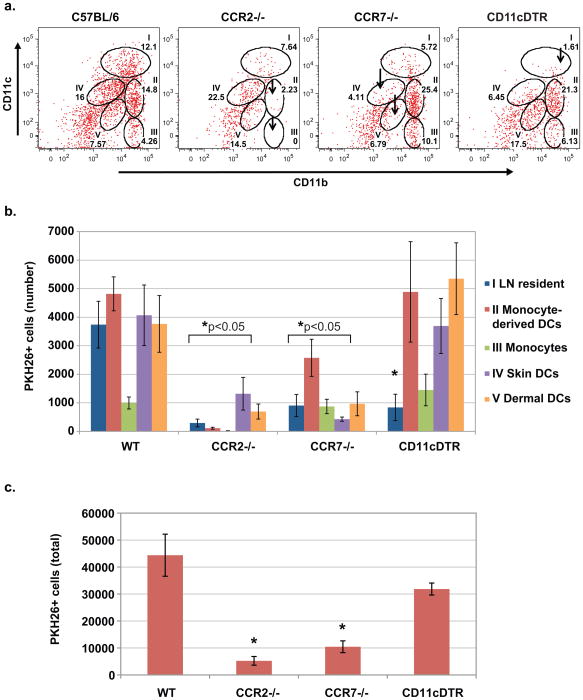
We concluded above that monocyte-derived DCs and skin-derived DCs are potential migratory populations that ferry vaccine yeast into the LN for possible Ag transfer to resident conventional DCs (Fig 1b). We used the chemokine system to restrict the migration of DC subsets into the LN and analyze the impact on distribution and number of PKH26+ cells in the LNs of vaccinated mice. Chemokine receptor 2 (CCR2) governs the migration of Ly6C+ monocytes (Serbina et al., 2008), and chemokine receptor 7 (CCR7), the migration of skin-derived DCs (Ohl et al., 2004). Vaccination of CCR2−/− and CCR7−/− mice sharply altered the distribution and number of PKH26+ cells in the draining LNs. In CCR2−/− mice, monocyte-derived DCs and monocytes (populations II and III) were greatly diminished, shifting PKH26+ cells to skin DCs (IV and V) (Fig. 3a and 3b). In CCR7−/− mice, skin-derived dermal and Langerhans DCs were reduced, shifting PKH26+ cells to the monocyte populations. The total number of PKH26+ cells in the draining LNs of vaccinated mice was also reduced – by about 2- to 4-fold – in absence of CCR2 or CCR7 (Fig. 3c). The CCR2−/− mice show the largest reduction of PKH26+ cells in the draining LNs, underscoring their prominent role in trafficking vaccine yeast (Fig. 2).
Figure 3. Delivery of vaccine yeast into the LNs of CCR2−/−, CCR7−/− and CD11cDTR mice.
Wild-type, or CCR2−/−, CCR7−/− and CD11cDTR mice were vaccinated with 107 PKH26+ yeast, and the draining LNs were harvested 3 d later at the peak of cell influx to analyze the presence and distribution of PKH26+ yeast in LN cells. CD11cDTR mice were depleted of LN-resident DCs as in Methods. The percent (A) and absolute number (B) of PKH26+ yeast-bearing cells are shown for the gated populations of DCs described in Figs. 1 and 2. Arrows indicate missing cell populations. *, p<0.05 vs. wild-type mice, except for monocytes in CCR7−/− mice. (C) The total number of PKH26+ cells in the LNs of vaccinated mice. Error bars represent the mean ± SD of 4 mice/group. Data is representative of 3 independent experiments. *, p<0.05 vs. wild-type mice.
Our findings in CCR2−/− and CCR7−/− mice support the view that migratory, monocyte-derived DCs and skin DCs (populations II, III, IV and V) ferry vaccine yeast into the nodes of vaccinated mice. To address the contribution of LN-resident DCs (population I), which are CD11chi, we used CD11cDTR mice, conditionally depleting CD11chi cells by injection of diphtheria toxin (Probst et al., 2005). In vaccinated mice, depletion of CD11chi cells eliminated LN-resident DCs with PKH+ yeast (Fig. 3a and 3b), but did not significantly alter the total number of PKH26+ cells in the draining LNs (Fig. 3c). Thus, eliminating CD11chi cells did not blunt the delivery of yeast into the LNs, but instead redistributed yeast Ag within the node, again suggesting that vaccine Ag might be transferred within the node from migratory DCs to resident DCs (Allan et al., 2006).
DC subsets that process and display vaccine yeast Ag in the draining LNs
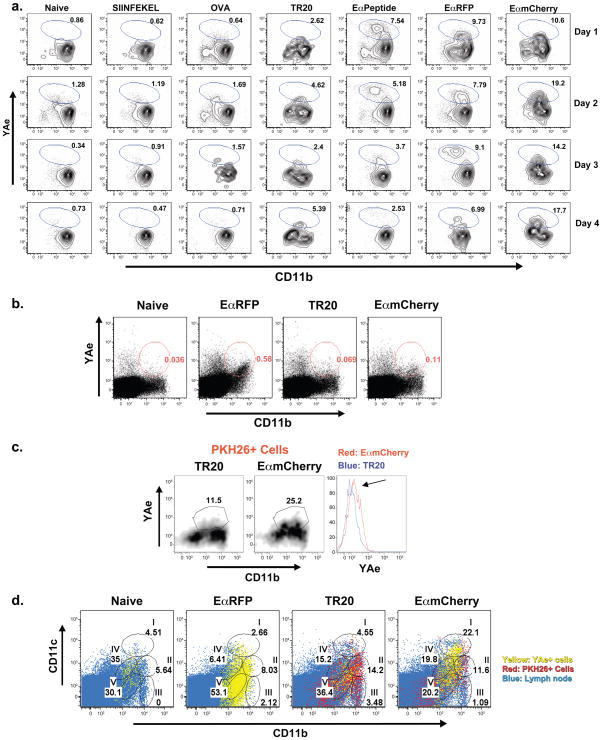
We observed that skin-derived DCs, monocyte-derived DCs, and LN-resident DCs contain vaccine yeast by PKH26+ staining, but the Ag-processing capability of these cells was not addressed. We used YAe antibody to detect Eα peptide displayed by MHCII (Itano et al., 2003), in conjunction with vaccine yeast engineered to express Eα (EαmCherry yeast), to identify APCs that display yeast-derived Ag. Since T-cell epitopes of B.dermatitidis are unknown, Eαserves as a derivative of the yeast surface protein BAD1 to let us monitor yeast-Ag display by APCs (see Methods). To validate this Ag detection system, we conducted in vitro studies with bone marrow derived dendritic cells (BMDCs). BMDCs co-cultured with products containing Eα sequence (Eαpeptide, soluble EαRFP and EαmCherry yeast) showed a distinct YAe+ population (Fig. 4a). Nearly 20% of the cells stained positive for Eα:MHCII complexes after co-culture from 1 to 4 days with EαmCherry yeast. In contrast, cells co-cultured with an irrelevant peptide (SIINFEKL), protein (OVA) or yeast lacking E α (TR20 control) showed low background staining with the YAe antibody (Fig. 4a).
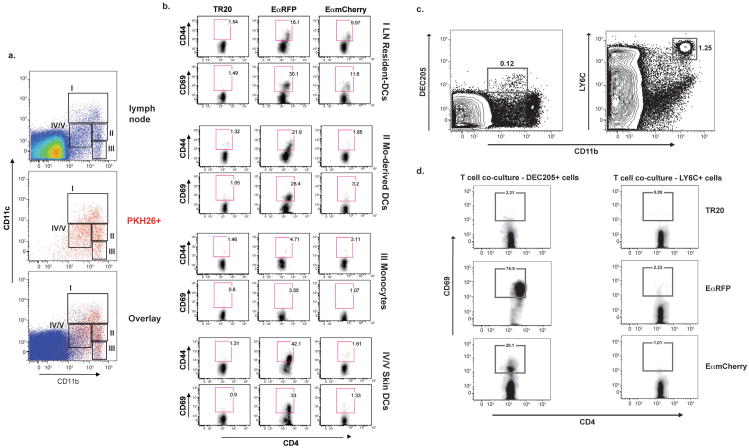
Figure 4. Surface display of vaccine yeast-derived peptide by APCs in the draining LNs.
(A) Utility of EαmCherry vaccine yeast and YAe Mab for detecting Eα:MHCII complexes on APCs. BMDCs (5×105 cells) were co-cultured in vitro with 10μM SINFEKEL peptide, 5μg/ml OVA, 5×105 TR20 yeast, 10μM Eαpeptide, 5μg/ml EαRFP, 5×105EαmCherry yeast (from left to right). Yeast were co-cultured with DCs for 16 hrs before non-adherent yeast were removed. Wells were harvested 1–4 d after co-culture for analysis of Eα:MHCII display detected by YAe Mab. (B) Detection of Eα:MHCII complexes on LN cells loaded in vivo. Mice were vaccinated once s.c. with 200 μg of EαRFP or with 107 PKH26+ TR20 yeast or Eαmcherry yeast. Cells from the draining LNs were analyzed with YAe mAb for EαMHCII display by FACS. (C) PKH26+CD11b+ cells from the LNs of vaccinated mice were gated on to analyze Eα: MHCII display by staining with YAe mAb. (D) Eα Ag display in vivo by cells in the LNs of vaccinated mice. Eα:MHCII display is depicted by overlaying YAe+ CD11b+ cells (yellow), PKH26+ cells (red) and total LN cells (blue). Mice received 200 μg soluble EαRFP or 107 PKH26+ TR20 yeast or EαmCherry yeast and LNs were harvested 3 d later. Cells were stained with 50–100μg YAe mAb. Numbers indicate percent of YAe+ cells in the gate for each population (as in Figs. 1 and 2). Results are representative of 3 separate experiments each with 3–4 mice/group.
To track cells that display vaccine yeast Eα Ag in vivo, we immunized mice with EαmCherry yeast or controls of TR20 yeast (negative) or soluble EαRFP (positive). The CD11b+ population of LN cells showed an increase in YAe+ staining in mice that got EαRFP or EαmCherry yeast, as compared to naïve mice and TR20 vaccine controls (Fig. 4b). CD11b+ cells from the EαmCherry yeast group had ≈2-fold more YAe+ cells than the TR20 group (Fig 4b), and gating on the CD11b+ PKH26+ LN cells accentuated the difference between these two groups (Fig. 4c). We also noted a constant population of YAe+CD11b− cells in all of our samples, consistent with Eα expression as a self-Ag (Murphy et al., 1992). Self-Ag display did not interfere with our identification of DCs that display yeast Ag since PKH26+ cells are CD11b+ rather than CD11b− (Fig. 1b). Thus, YAe staining together with gating on PKH26 and CD11b enabled us to discriminate the display of yeast E αfrom self Ag.
To identify the APCs that process and display yeast-derived Eα in relation to the Ag-loaded cells, we overlaid the LN populations of YAe+ cells and PKH26+ cells for mice that were vaccinated with EαRFP, TR20 yeast, or EαmCherry yeast. In the EαRFP (soluble protein)-vaccinated animals, the majority of YAe+ cells reside within the skin-derived DC populations IV and V (Fig. 4d), similar to what has been reported (Itano et al., 2003). In contrast, the most abundant display of Eα:MHCII in EαmCherry yeast-vaccinated animals occurs within the LN-resident conventional DCs, with surprisingly few YAe+ cells in the monocyte-derived DC and monocyte/macrophage gates (Fig. 4d). This finding implies that monocyte-derived cells carry yeast into the LN, but may play a limited role in directly displaying yeast Ag to naïve T cells. Because Eα:MHCII display is necessary but not sufficient for Ag presentation to naïve T cells, we sought to identify the DCs capable of both displaying yeast Ag and priming naïve Ag-specific T cells in the LN.
Naïve TEa T cells report fungal Ag processing and presentation in the draining LNs
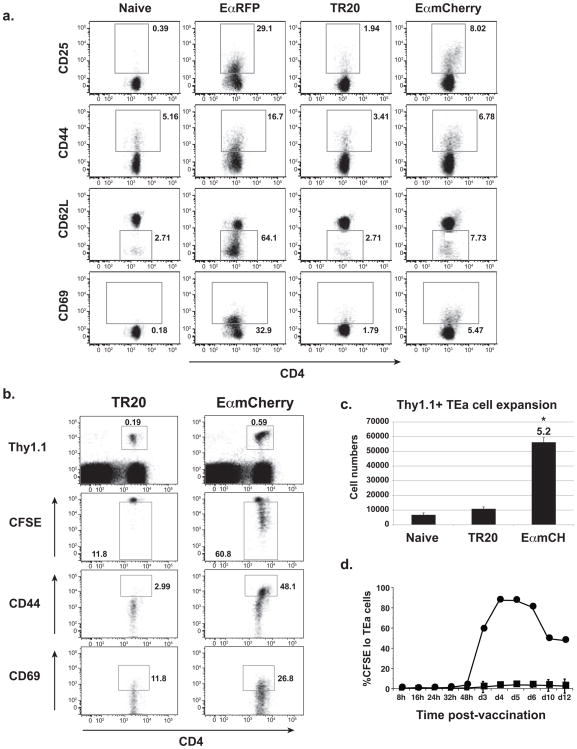
Using naïve TEa Tg T cells specific for Eα:MHCII complexes (Murphy et al., 1992; Rudensky et al., 1991), we determined how the display of fungal Eα Ag in the draining nodes, as detected by the YAe antibody, correlates with the priming of naïve T cells. We found above that CD11b+ cells in the LN associate with vaccine yeast (Fig. 1b) and display yeast-derived Eα Ag (Fig. 4b). We therefore collected CD11b+ cells from mice vaccinated with soluble EαRFP, EαmCherry yeast, or TR20 control yeast and co-cultured them ex vivo with TEa cells to see if Ag display activated the naïve T cells. CD11b+ cells from the soluble EαRFP and EαmCherry yeast groups were able to activate the TEa cells as observed by increased surface expression of CD25, CD44, CD69, and decreased expression of CD62L (Fig 5a); CD11b+ cells from naïve mice and TR20 controls, and CD11b− cells from any vaccine group failed to activate the TEa cells (Fig 5a, and data not shown).
Figure 5. Naïve TEa cells report fungal Ag presentation in vivo by responding to Eα:MHCII.
(A) TEa T cells report yeast Eα-Ag presentation ex vivo by cells from the draining LNs of immunized mice. CD11b+ cells (105) from naïve mice or those vaccinated with soluble EαRFP, or TR20 (control) yeast or EαmCherry yeast were harvested 3 d after vaccination and cultured ex vivo with 5×105 naïve CD4+ TEa T-cells. After 3 d of co-culture, TEa cells were stained for activation markers. Data are representative of 3 independent experiments with 3–4 mice/group. (B) Naïve TEa cells report vaccine yeast Eα-Ag presentation in vivo. Thy 1.1 TEa T cells were labeled with CFSE and transferred into C57BL/6 mice (105 cells/mouse) vaccinated with 106 TR20 control or Eαmcherry yeast. Draining LNs were collected 4 d later, stained for activation markers and analyzed by FACS. T cells were gated on Thy1.1+ TEa cells. Numbers are percent of cells in the gate. Data are representative of 3 experiments with 4 mice/group. (C) Fold-expansion (numbers) of TEa cells was quantified for the experiment in panel B. *p<0.05 vs. TR20 or naïve mice. (D) The dynamics of vaccine Eα presentation to T-cells in vivo. Thy1.1 TEa cells were labeled with CFSE and transferred into mice. After transfer, mice received 106 TR20 control (square) or EαmCherry yeast (circle) s.c. Draining LNs were analyzed serially to assay proliferation (CFSElo) of Thy1.1+ TEa cells. The data are the mean ± SD of 3 independent experiments with 5 mice/group at each time point.
To see if vaccine Eα display in the LN primed naïve TEa cells in vivo, we adoptively transferred TEa cells into mice and vaccinated them with EαmCherry or TR20 control yeast (Fig. 5b). Three days after vaccination, TEa cells in the EαmCherry-vaccinated animals were primed, as noted by their CFSElo and CD44hi and CD69hi activated phenotype (Fig. 5b). In the TR20 group, in contrast, TEa cells failed to become activated and retained the naïve phenotype of CFSEhi, CD44lo and CD69lo. Thus, although staining of draining LNs with antibody showed only a small increase in the YAe+ population in mice vaccinated with EαmCherry yeast vs. TR20 yeast (Fig. 4c), the population is biologically active in priming naïve Ag-specific T cells. Eα displayed as a self-Ag in the CD11b− population in the LN (Figs. 4b and 4c) did not interfere with the specificity of TEa cells for vaccine yeast-derived EαAg since naïve TEa cells failed to become activated in either unvaccinated mice or those that received TR20 control yeast (Fig. 5b). Naïve TEa cells expanded 5- to 8-fold more in mice vaccinated with EαmCherry compared to these control groups (Fig. 5c).
Soluble Ag administered s.c. enters the LN in two waves, at 30 minutes and again at ~14 hours, with corresponding early T-cell priming (Itano et al., 2003). We observed that vaccine yeast are dynamically distributed among APC populations in the draining LNs of mice over 7 days after vaccine administration. Here, we analyzed how the kinetics of vaccine Ag delivery into the draining nodes correlates with the kinetics of priming naïve T cells at this site. After adoptive transfer of TEa cells, their activation and proliferation was monitored for 12 days in response to vaccination with EαmCherry or control yeast. TEa cells initially proliferated at ≈72 hours in response EαmCherry yeast, as measured by their loss of CFSE (Fig. 5d). Analysis of other T cell activation markers, such as surface CD25, CD44 or CD69, showed evidence of TEa activation about 24–36 hours earlier (data not shown). Thus, priming of naïve T cells by vaccine yeast correlates closely with the initial appearance of PKH26+ signal in the draining LNs of vaccinated mice (Fig. 1a). The lack of evidence of T cell priming before ~32 hours in response to vaccine yeast supports the idea that fungal Ag is actively ferried by cells into the draining LN and does not enter by passive diffusion through the lymphatics (Iezzi et al., 2006; Itano et al., 2003). Although the soluble vaccine Ag E αRFP primed TEa cells sooner, yeast particulate Ag induced 3-fold more expansion of TEa cells at the peak of T cell priming. Our findings also show that the system here of TEa cells and EαmCherry yeast should let us investigate and identify the DC subsets that prime naïve T cells in response to fungal vaccination.
DC subsets from vaccinated mice prime naïve T-cells ex vivo
Because we identified three subsets of CD11b+CD11c+ DCs (skin-derived DCs, LN-resident DCs, and monocyte-derived DCs) associated with vaccine yeast in the LN, we determined whether any or all of these DC subsets may be responsible for priming protective T cells in this model of immunization against fungi. Based on the kinetics of T cell priming in vivo, which showed initial T cell expansion at 72 hours in response to EαmCherry yeast (Fig. 5c, Fig. 5d), we collected and functionally analyzed in vivo loaded APCs from the draining LNs three days after vaccination. The cells were sorted by FACS according to gates that correspond to the different DC subsets (Fig. 6a). Four populations were sorted from total LN cells: resident DCs (population I), monocyte-derived inflammatory DCs (II), monocytes/macrophages (III) (as a negative control), and skin-derived dermal and Langerhans DCs (IV) (Fig. 6a). Populations from mice vaccinated with soluble EαRFP, EαmCherry yeast or TR20 control yeast were sorted in parallel. TEa cells were co-culturedwith sorted cells ex vivo for 72 hours and analyzed for the expression of activation markers. In the EαmCherry yeast-vaccine group, LN-resident DCs activated the TEa cells, as defined by their increased expression of surface CD44 and CD69, when compared to the TR20 control group; no other sorted populations from the EαmCherry group activated the TEa cells (Fig. 6b). In the soluble EαRFP group, skin DCs activated the TEa cells, as described (Itano et al., 2003), although monocyte-derived DCs and LN-resident DCs were also capable of activating the cells in this ex vivo assay.
Figure 6. The identity of APCs that prime T cells in response to fungal vaccine.
Draining LN cells were sorted by FACS to identify DC subsets that prime naïve TEa T cells ex vivo after immunization. (A) 3 d after vaccination with 100μg EαRFP, or 107 heat-killed TR20 control yeast or EαmCherry yeast, LN cells were harvested from 2–3 mice/group. Gates were based upon FSC and SSC, then FSC-W for singlet cells, and finally on B220− cells. CD11b+ cells were divided into 4 populations for sorting. (B) Sorted population were cultured (2×104 cells) ex vivo with bead-enriched CD4+ naïve TEa cells (3×105) for 72 hrs, and the cultures harvested and stained for T cell activation markers. Roman numerals and names correspond to sorted populations in panel A. Numbers in gates are the percent of TEa cells with the activation marker. Results are representative of 3 independent experiments pooling LNs of 2 mice/group. (C) Skin-derived DCs sorted from the LNs of vaccinated mice. DEC205 (left panel) was used to sort skin-derived DCs, and LY6C (right panel), to collect cells as a “negative” control for culture ex vivo with TEa cells in panel D. Numbers reflect percent of cells in the gate. (D) Sorted DEC205+ cells and control LY6C+ cells (as in panel C) from vaccinated mice were cultured (2×104) ex vivo with bead-enriched naïve TEa cells (3×105). After 72 hrs, T cells were stained for activation and analyzed by FACS. Results are representative of 2 independent experiments with 2 mice/group.
We were surprised that skin DCs (population IV in Fig. 6a) did not prime TEa cells in EαmCherry-vaccinated mice since the fungal vaccine is most effective when injected s.c. (Wüthrich et al., 2000). One possibility is that skin DCs were diluted by other cell types since population IV is sorted from total LN cells. We observed in Fig. 1c that populations IV and V are heterogeneous, and only a minor proportion of them are DEC205+ skin DCs (~20% of the PKH26+ population). This raised the possibility that the lack of T cell activation with this population was due to a non-optimal ratio of DCs to TEa cells in vitro. To address this, we sorted DEC205+ cells in vaccinated mice to enrich skin DCs (Fig. 6c), and in parallel we sorted Ly6C+ monocytes as a control, which did not prime TEa cells in the EαmCherry group (Fig 6b). We obtained 10-fold less cells by sorting DEC205+ cells compared to sorting population IV in Fig. 6a, suggesting that skin DCs were indeed “diluted out” by other LN cells in the original gating strategy. DEC205+ cells loaded in vivo with EαmCherry yeast activated TEa cells following co-culture. Over 20% of TEa cells expressed surface CD69 in this vaccine group, compared to ≈3% of cells in the TR20 control group. DEC205+ skin DCs loaded in vivo with soluble EαRFP also primed naïve TEa cells as expected (Itano et al., 2003). Thus, skin-derived DCs from mice vaccinated with yeast can also prime naïve T cells ex vivo. The Ly6C+ sorted cells contained 5-fold more cells than population II because the sorted cells are composed of multiple monocyte-derived populations including monocytes, monocyte-derived DCs and macrophages. However, the Ly6C+ cells from each vaccine group failed to prime TEa cells.
Analysis of T cell priming in vivo unveils collaborative, indispensable roles of DC subsets
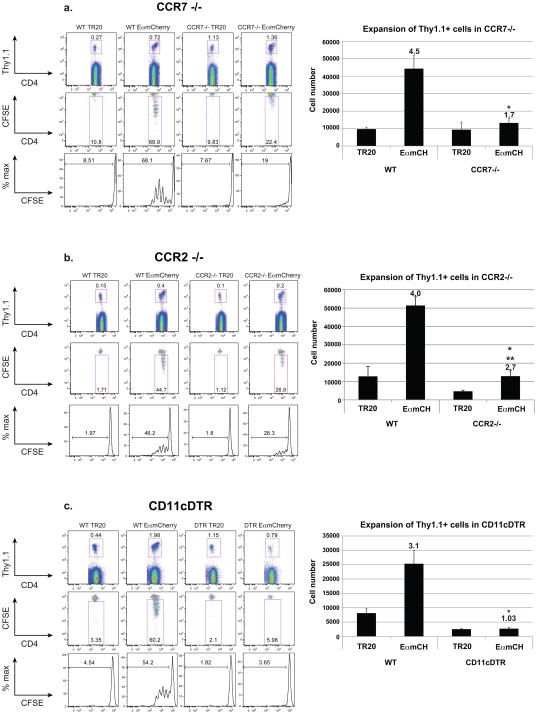
Ex vivo assays do not necessarily portray the functional behavior of APCs in vivo, where distinct DC subsets may collaborate to prime naïve T cells (Allenspach et al., 2008). Hence, we studied the in vivo role of distinct DC subsets in priming naïve T cells in response to vaccination. We utilized CCR2−/−, CCR7−/− and CD11cDTR mice (Fig. 7a–c). We adoptively transferred naïve TEa cells into these mice and monitored T-cell activation after vaccination with EαmCherry or TR20 control yeast. Our sort data above revealed that LN-resident DCs and skin DCs loaded with Ag in vivo can prime naïve T cells ex vivo (Fig. 6). The role of these DC subsets in in vivo priming was confirmed in CCR7−/− and CD11cDTR mice (Fig. 7a and c). CCR7−/− mice, in which migration of skin DCs is impaired, showed sharply reduced priming of naïve TEa cells despite our earlier finding that skin DCs were a minor (<20%) yeast-associated population in the draining LNs (Fig. 1b). Nearly 69% of naïve TEa cells were CFSElo and primed in wild-type mice vaccinated with EαmCherry yeast, whereas 22% of the cells were primed in CCR7−/− mice (Fig. 7a). Impairment in TEa priming was even more striking in CD11cDTR mice, which lack conventional, LN-resident DCs, and where the absence of CD11chi cells virtually abrogated TEa cell proliferation (Fig. 7a).
Figure 7. Impairments in vaccine priming of T cells after the loss of DC populations.
In vivo priming of Ag specific T-cells. (A) CCR7−/−, (B) CCR2−/− and (C) CD11cDTR mice were used to analyze T-cell priming in the absence of skin-derived DCs, monocyte-derived DCs and LN-resident DCs, respectively. Naïve Thy1.1+ TEa T cells (2×105) were labeled with CFSE and transferred into recipients a day before vaccination s.c. with TR20 control yeast or EαmCherry yeast (107). Draining LNs were harvested 3 d after vaccination and analyzed by FACS. Thy1.1+ cells (TEa cells) were gated (upper row) and analyzed for proliferation in vivo. Percent of cells proliferating is displayed within each gate (2nd row) or histogram (3rd row). Numbers and fold-expansion of Thy1.1+ TEa cells is shown on the right. Numbers are fold-change vs. TR20 control. *p<0.05 for EαmCH in WT vs. transgenic strain. **p<0.05 for EαmCH vs. TR20. The results are representative of 3 independent experiments with 3 mice/group. Results were similar when LNs were harvested 48 hrs after vaccination (Fig. S7).
Similarly, the priming of TEa cells was partially impaired in CCR2−/− mice; proliferation of TEa cells in these mice reached ≈28% of the maximal response, as compared to 46% in wild-type mice (Fig. 7b). Hence, although monocyte-derived inflammatory DCs are the least efficient subset in displaying Eα:MHCII after Ag-loading in vivo, and our ex vivo analysis of sorted monocyte-derived inflammatory DCs demonstrated that they are incapable of priming TEa cells in vitro, these DCs have a requisite role in the priming of anti-fungal T cells in vivo in response to vaccination. This is perhaps not surprising because monocyte-derived DCs are the first to infiltrate the LNs with vaccine yeast after immunization and this population contains the majority of Ag-loaded cells (Fig. 1b). Thus, while ex vivo analysis pointed to the functional role of two of the three DC subsets – skin DCs and lymph node DCs, but not monocyte-derived DCs in priming TEa cells – in vivo studies establish that the various DC subsets mediate distinct, collaborative and indispensable roles in priming T cells in response to vaccine.
DISCUSSION
We provide here a comprehensive picture of the earliest stages in the generation of vaccine immunity to a systemic fungal infection. Because DCs are essential in understanding how vaccine immunity is generated (Steinman, 2008), we focused on elucidating their role in our model. We found that multiple DC subsets collaborate to prime naïve Ag-specific CD4 T cells, and that the different subsets mediate distinct functions. Monocyte-derived inflammatory DCs mainly transport vaccine yeast Ag into the LN, whereas LN-resident DCs and, to a lesser extent migrating skin DCs, directly prime naïve T cells. Our work reveals an intricate interplay between DC subsets during the generation of vaccine immunity to fungi.
Monocyte-derived inflammatory DCs are the earliest and largest cell population that associates with vaccine yeast in the LN. We identified monocyte-derived DCs by their Ly6Chi CD11cint phenotype (Leon and Ardavin, 2008; Nakano et al., 2009). These cells were not detected in naïve mice, and their numbers increased sharply after vaccination, supporting their characterization as inflammatory cells. In CCR2−/− mice, their recruitment into the draining LN was curtailed, and the delivery of vaccine yeast was also reduced. While these DCs played a key role in early recognition, uptake and trafficking of yeast into the LN, they did not efficiently process and present Ag or prime naïve T cells. For such analyses, we exploited the TEa system. Vaccine yeast expressing EαRFP were used to track yeast-derived Eα:MHCII complexes on APCs with YAe mAb, and monitor priming of naïve Eα-specific “anti-fungal” TEa cells. Monocyte derived DCs had little processed Eα on their surfaces. In vaccinated mice, these cells were unable to prime naïve TEa cells ex vivo.
This discrete role of monocyte-derived DCs in trafficking vaccine yeast to the draining node contrasts with their broader functions in other eukaryotic models. In a L. major cutaneous infection model, Leon et al. (Leon et al., 2007) detected two monocyte-derived inflammatory DC populations: lymph node monocyte-derived DCs (LN mo-DC) are an immature Ly6Chi population, and dermal monocyte-derived DCs (dermal mo-DCs) are a more mature Ly6int population. The dermal mo-DCs were found to be the principal population expressing L. major LACK Ag peptide, and activating Ag-specific T cells ex vivo.
In our system, the inflammatory DC is phenotypically most similar to LN mo-DCs (Ly6ChiCD11bhiCD11cintMHCIIlo) described by Leon et al. (Leon et al., 2007). Those cells are purportedly monocytes that enter the LN directly from the blood and differentiate into DCs. There, DCs would be located along high endothelial venules and sample soluble Ags that enter via the lymph. The monocyte-derived inflammatory DCs in our model must be different. They are the first cells to associate with yeast in the LN (24 hours after vaccination). However, these monocyte-derived DCs don’t acquire PKH26+ yeast Ag via the lymph. Soluble Ag typically enters the node by 30 minutes after injection into the skin; DCs that ferry Ag enter later, by around 14 hours after injection (Iezzi et al., 2006; Itano et al., 2003). In mice vaccinated with yeast, we first detected PKH+ signal in the node at 24 hours; therefore it was likely ferried by DCs. After fungal vaccine, proliferation of naïve Eα specific TEa cells in vivo was delayed until 72 hours, also consistent with cell transport of Ag into the node. Third, impaired monocyte migration in CCR2−/− mice delayed the kinetics and amount of PKH26+ signal in the draining LN, demonstrating that these monocyte-derived DCs transport yeast into the node from the site of vaccination rather than sampling soluble, cell free Ag that arrives via the lymph.
The other subset of monocyte-derived inflammatory DCs in the L. major model were dermal mo-DCs (Ly6Clo, CD11bint, CD11cint MHCIIhi) (Leon et al., 2007). Neither does our inflammatory DC match that phenotype. Our cells may be an early phenotype of the dermal-mo DCs, since that subset was described at 4 weeks after infection with L. major. Our model instead focused on the initial stages of vaccination at 3 days post injection. Thus, yeast-associated monocyte-derived DCs are neither dermal mo-DC, nor LN mo-DCs reported by Leon et al. (Leon et al., 2007), but likely another monocyte population that arrives at the skin, quickly picks up vaccine yeast, differentiates into CD11b+ DCs, and transports Ag into the LN. Differences between the two studies may be due to the differences in timing of analysis or burden of Ag at those points during infection, and especially the differing nature of these eukaryotic pathogens.
The Ly6Chi monocyte-derived DC subset in our model is similar in phenotype and kinetics to inflammatory monocyte-derived CD11b+ DCs in models of pulmonary aspergillosis (Hohl et al., 2009) and crytococcosis (Osterholzer et al., 2009). Hohl et al. reported that Ly6ChiCD11b+CD11c+ DCs harbored conidia in the lung 2 days post-infection. These DCs required CCR2 for recruitment into the lung and draining LNs where they induced Th1 differentiation of naïve Aspergillus-specific Tg T cells. We also saw that loss of CCR2 delayed the delivery of vaccine yeast into the draining LNs and reduced the numbers of APCs associated with yeast. In contrast, however, we found that loss of CCR2 only partially impaired priming of naïve Ag specific T cells. The Aspergillus work did not address how monocyte-derived DC contribute to lung T cell priming; that is, whether the DCs solely transported Ag to the node, primed T cells or both. Nevertheless, the precise function(s) of monocyte-derived DCs were shown to differ with the tissue site. DCs were essential in priming T-cells in the lung, but dispensable for systemic priming in the spleen.
By using YAe antibody to detect Ag display, we went beyond the detection of cell Ag load by PKH26 signal and monitored the ability of distinct DC subsets to display yeast-derived peptide on their surfaces. Importantly, resident DCs displayed more surface peptide than migratory DC subsets. In CCR2−/− and CCR7−/− mice, however, PKH26+ signal and surface peptide display were reduced in resident DCs, suggesting that yeast Ag is transferred from migratory DCs (skin- or monocyte-derived) to resident DCs. YAe staining also revealed that different DC subsets presented soluble protein (EαRFP) versus live yeast. Whereas Ag display after soluble EαRFP injection was seen in the skin-derived DC subsets, vaccine yeast triggered greater display of processed Ag in the LN-resident DCs. Although monocyte-derived DCs contained the largest burden of PKH+ yeast, they displayed the fewest Eα:MHCII complexes. Thus, engulfing many yeast may have impaired Ag presentation in these cells. In a Herpes Simplex Virus-1 (HSV-1) model, the large viral Ag load impaired DC function and was associated with defective cell trafficking (Eidsmo et al., 2009).
Based on YAe staining and Ag display, we postulated that mainly skin DCs, after soluble EαRFP vaccination, and LN-resident DCs, after vaccine yeast administration, would prime naïve T cells. Our ex vivo T cell assays with sorted cells confirmed the YAe staining results and established that in vivo-loaded LN-resident DCs and, to a lesser extent, migratory skin-derived DCs, primed naïve T cells. However, previous studies in an OVA-OT II system showed that migratory and LN-resident DCs could prime T cells ex vivo independently of each other, but in vivo priming required the cooperation of both subsets (Allenspach et al., 2008). In keeping with that study, we observed reduced priming of naïve T cells in CD11cDTR mice (lack LN-resident DCs [CD11chi cells]) (Bennett and Clausen, 2007) and CCR7−/− mice (impaired entry into the LN of migratory DCs) (Jang et al., 2006; Ohl et al., 2004). In CD11cDTR mice, T cells were not primed in vivo; in CCR7−/− mice, T cell priming was reduced ~3-fold. Any residual response in CCR7−/− mice might be due to monocyte-derived DCs that compensated for skin (likely dermal) DCs as the migratory subset. Although monocyte-derived DCs are not efficient in priming T cells directly, they likely transfer Ag from the skin to LN-resident cells, thereby preserving T cell priming in CCR7−/− mice. Our CCR7−/− results contrast with those in CCR2−/− mice, where the defect in T cell priming was less pronounced. There, the putative role of CCR2+ monocyte-derived DCs likely could be rescued partially by other populations in the skin, likely the dermal DCs. This finding reinforces the conclusion by Hohl et al. (Hohl et al., 2009) that tissue site influences the role of monocyte derived DCs in response to fungi. Our study offers new insight into how these DCs help prime T-cells to yeast injected into the skin, and promote vaccine immunity to fungi.
Lymph node-resident DCs were indispensable in the generation of vaccine immunity to fungi, in our model. This DC subset made up the bulk of surface Ag-displaying cells in the draining LNs of vaccinated mice, and they primed naïve TEa T cells ex vivo, and were required in priming them in vivo. Lymph node resident DCs also have a seminal role in inducing T cell immunity against Leishmania and HSV-1 (Allan et al., 2006; Iezzi et al., 2006). In HSV-1 infection, the route of inoculation determines which DC subset induces the activation of T cells (Lee et al., 2009). Dermal DCs prime T cells when HSV is introduced via the mucosa, whereas LN-resident DCs show a more prominent role after needle inoculation (Lee et al., 2009). One explanation for the effectiveness of our fungal vaccine when given s.c., may be that the LN-resident DCs in skin draining LNs are available to induce protective immunity. Conversely, tissue DCs at other routes or sites of infection may not be as efficient in priming anti-fungal T cells. This may explain the low efficacy of our fungal vaccine given by the respiratory route (Wüthrich et al., 2000).
Most studies of the role of DCs in priming antimicrobial T cell responses have emphasized the seminal role of a distinct DC subset, according to the pathogen or the site of entry or injection. In contrast, our study reveals the interplay among multiple, distinct subsets. Whereas both monocyte-derived and dermal DCs ferry vaccine yeast into the draining LN, neither subset fully compensates for the other in chemokine receptor-deficient mice, and the priming of CD4 T cells was impaired cells in both CCR2−/− and CCR7−/− mice. CCR7−/− mice showed a striking defect in T cell priming, likely due to the multiple functions of dermal DCs: trafficking and transferring of Ag to resident DCs, as well as priming of T cells. We did not formally show Ag transfer to resident DCs, but the findings that Ag must be ferried into the node by cell migrants, and that blocking cell migration by toxin 24 hours after vaccination failed to curtail the uptake of Ag in resident DCs is most consistent with that mechanism.
Our study exploited a model Ag to clarify sequential steps in initiating Ag-specific anti-fungal immunity, but it is valid to ask whether the response to this model Ag accurately portrays vaccine immunity to fungi. We recently used CD4 T-cells from a newly created Blastomyces-specific TCR tg mouse, which confirmed our results here with TEa cells (unpublished data). Overall, our results typify the cooperation of multiple DC subsets needed to prime CD4 T cells in response to the model Ag ovalbumin (Allenspach et al., 2008). Upon s.c. injection of live attenuated yeast, the generation of vaccine immunity likewise demands the dynamic and functional collaboration of both migratory and resident DC subsets.
MATERIALS AND METHODS
Mice
Wild-type C57BL/6 mice were obtained from the National Cancer Institute, Bethesda MD. TEa TCR transgenic mice were provided by A.Y. Rudensky at the Howard Hughes Medical Institute, University of Washington (Itano et al., 2003). CCR2−/− (stock#04999), CCR7−/− (stock# 05794) and CD11cDTR/GFP mice (stock# 04509) were obtained from Jackson Laboratories. CD11chi cells were depleted from CD11cDTR/GFP mice by one intra-peritoneal injection of 100ng of diphtheria toxin per animal. Mice were 10–12 weeks of age at the time of our experiments. Experimental procedures and housing were performed according to the guidelines of the University of Wisconsin Animal Care Committee, who approved this work.
Fungi
Blastomyces strain 55 is an attenuated yeast created from the isogenic virulent strain American Type Culture Collection (ATCC) 26199 (ATCC, Manassas, VA) by targeting and deleting the gene encoding BAD1 (Brandhorst et al., 1999). EαmCherry yeast was engineered as described in Supplemental Methods. Strain TR20 served as a control in some experiments. All fungal isolates were maintained in the yeast form at 39°C on Middlebrook 7H10 agar slants.
Staining of vaccine yeast and subcutaneous immunization
Yeasts were labeled with the red-fluorescent linker PKH26GL (Sigma, St. Louis, MO) in Diluent C (Sigma) at a concentration of 2 × 106 yeast/μl PKH26 for 5 minutes at room temperature. An equal volume of FBS (Cell Gro®, Mediatech Inc., Manassas) was added to bind excess PKH26 for one minute, followed by a single wash with RPMI medium supplemented with 10% FBS, and then by two washes with PBS. Yeasts were labeled with CFSE using the manufacturer’s instructions (Molecular Probes) for lymphocytes, which yields strong fluorescent intensity. For vaccination, PKH-labeled yeasts were suspended at 1×107 cells/ml in PBS, and two injections were given s.c. Each injection site received 5×106 yeast in 500μl. Approximately100–200μg of EαRFP in PBS was administered s.c. as for vaccine yeast. The EαRFP (Eα plus DSRed) protein was expressed in recombinant form in Escherichia coli and purified as described by Jenkins et al. (Itano et al., 2003). DC migration into the draining LN was blocked by adding 500ng of pertussis toxin (Sigma-Aldrich) to vaccine yeast during s.c. injection. To inhibit DC migration following vaccination, 300ng of pertussis toxin was given i.v. 24 hours after vaccine yeast were injected.
T Cell preparation and adoptive transfers
Naïve CD4+ T-cells were harvested from the spleen and LNs of naïve TEa mice. Splenocytes and LN single-cell suspensions were enriched on CD4 magnetic beads (IMAG, BD Bioscience). T cells were enriched as per the manufacturer’s instructions. T cells were co-cultured with various concentrations of DCs at 3×105 T cells/well. For adoptive transfers, single cell suspensions of whole splenocytes of naïve TEa Thy1.1+ mice were stained with CFSE (Molecular Probes) before transfer as per the manufacturer’s instructions. CFSE-labeled TEa cells were transferred intravenously (i.v.) at 106 or 2×106 per animal to obtain ~105 to 2×105 CD4+ T cells respectively, since ≈10% of total splenocytes are CD4+ TEa T cells. A day after transfer mice were vaccinated with 106 yeast s.c., and brachial and inguinal LNs were harvested 3 days later.
Statistics
Differences in the number of cells were analyzed using Wilcoxon rank test for non-parametric data or a T-test if data were normally distributed. A Bonferroni adjustment was used to correct for multiple tests. A value of P < 0.05 is considered significant.
Supplementary Material
Acknowledgments
This work was supported by the USPHS. Nicole Rocco provided comments on the manuscript. Robert Gordon assisted with illustrations.
References
- Allan RS, Waithman J, Bedoui S, Jones CM, Villadangos JA, Zhan Y, Lew AM, Shortman K, Heath WR, Carbone FR. Migratory dendritic cells transfer antigen to a lymph node-resident dendritic cell population for efficient CTL priming. Immunity. 2006;25:153–162. doi: 10.1016/j.immuni.2006.04.017. [DOI] [PubMed] [Google Scholar]
- Allenspach EJ, Lemos MP, Porrett PM, Turka LA, Laufer TM. Migratory and lymphoid-resident dendritic cells cooperate to efficiently prime naive CD4 T cells. Immunity. 2008;29:795–806. doi: 10.1016/j.immuni.2008.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banchereau J, Steinman RM. Dendritic cells and the control of immunity. Nature. 1998;392:245–252. doi: 10.1038/32588. [DOI] [PubMed] [Google Scholar]
- Bennett CL, Clausen BE. DC ablation in mice: promises, pitfalls, and challenges. Trends Immunol. 2007;28:525–531. doi: 10.1016/j.it.2007.08.011. [DOI] [PubMed] [Google Scholar]
- Bradsher RW. Systemic fungal infections: diagnosis and treatment. I. Blastomycosis. Infectious Disease Clinics of North America. 1988;2:877–898. [PubMed] [Google Scholar]
- Brandhorst TT, Wüthrich M, Warner T, Klein B. Targeted gene disruption reveals an adhesin indispensable for pathogenicity of Blastomyces dermatitidis. J Exp Med. 1999;189:1207–1216. doi: 10.1084/jem.189.8.1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown GD. Dectin-1: a signalling non-TLR pattern-recognition receptor. Nat Rev Immunol. 2006;6:33–43. doi: 10.1038/nri1745. [DOI] [PubMed] [Google Scholar]
- Dudziak D, Kamphorst AO, Heidkamp GF, Buchholz VR, Trumpfheller C, Yamazaki S, Cheong C, Liu K, Lee HW, Park CG, et al. Differential antigen processing by dendritic cell subsets in vivo. Science. 2007;315:107–111. doi: 10.1126/science.1136080. [DOI] [PubMed] [Google Scholar]
- Eidsmo L, Allan R, Caminschi I, van Rooijen N, Heath WR, Carbone FR. Differential migration of epidermal and dermal dendritic cells during skin infection. J Immunol. 2009;182:3165–3172. doi: 10.4049/jimmunol.0802950. [DOI] [PubMed] [Google Scholar]
- Heath WR, Belz GT, Behrens GM, Smith CM, Forehan SP, Parish IA, Davey GM, Wilson NS, Carbone FR, Villadangos JA. Cross-presentation, dendritic cell subsets, and the generation of immunity to cellular antigens. Immunol Rev. 2004;199:9–26. doi: 10.1111/j.0105-2896.2004.00142.x. [DOI] [PubMed] [Google Scholar]
- Hohl TM, Rivera A, Lipuma L, Gallegos A, Shi C, Mack M, Pamer EG. Inflammatory monocytes facilitate adaptive CD4 T cell responses during respiratory fungal infection. Cell Host Microbe. 2009;6:470–481. doi: 10.1016/j.chom.2009.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iezzi G, Frohlich A, Ernst B, Ampenberger F, Saeland S, Glaichenhaus N, Kopf M. Lymph node resident rather than skin-derived dendritic cells initiate specific T cell responses after Leishmania major infection. J Immunol. 2006;177:1250–1256. doi: 10.4049/jimmunol.177.2.1250. [DOI] [PubMed] [Google Scholar]
- Itano AA, McSorley SJ, Reinhardt RL, Ehst BD, Ingulli E, Rudensky AY, Jenkins MK. Distinct dendritic cell populations sequentially present antigen to CD4 T cells and stimulate different aspects of cell-mediated immunity. Immunity. 2003;19:47–57. doi: 10.1016/s1074-7613(03)00175-4. [DOI] [PubMed] [Google Scholar]
- Jang MH, Sougawa N, Tanaka T, Hirata T, Hiroi T, Tohya K, Guo Z, Umemoto E, Ebisuno Y, Yang BG, et al. CCR7 is critically important for migration of dendritic cells in intestinal lamina propria to mesenteric lymph nodes. J Immunol. 2006;176:803–810. doi: 10.4049/jimmunol.176.2.803. [DOI] [PubMed] [Google Scholar]
- Kaplan DH, Jenison MC, Saeland S, Shlomchik WD, Shlomchik MJ. Epidermal langerhans cell-deficient mice develop enhanced contact hypersensitivity. Immunity. 2005;23:611–620. doi: 10.1016/j.immuni.2005.10.008. [DOI] [PubMed] [Google Scholar]
- Kaplan DH, Kissenpfennig A, Clausen BE. Insights into Langerhans cell function from Langerhans cell ablation models. Eur J Immunol. 2008;38:2369–2376. doi: 10.1002/eji.200838397. [DOI] [PubMed] [Google Scholar]
- Lee HK, Zamora M, Linehan MM, Iijima N, Gonzalez D, Haberman A, Iwasaki A. Differential roles of migratory and resident DCs in T cell priming after mucosal or skin HSV-1 infection. J Exp Med. 2009;206:359–370. doi: 10.1084/jem.20080601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leon B, Ardavin C. Monocyte migration to inflamed skin and lymph nodes is differentially controlled by L-selectin and PSGL-1. Blood. 2008;111:3126–3130. doi: 10.1182/blood-2007-07-100610. [DOI] [PubMed] [Google Scholar]
- Leon B, Lopez-Bravo M, Ardavin C. Monocyte-derived dendritic cells formed at the infection site control the induction of protective T helper 1 responses against Leishmania. Immunity. 2007;26:519–531. doi: 10.1016/j.immuni.2007.01.017. [DOI] [PubMed] [Google Scholar]
- Lopez-Bravo M, Ardavin C. In vivo induction of immune responses to pathogens by conventional dendritic cells. Immunity. 2008;29:343–351. doi: 10.1016/j.immuni.2008.08.008. [DOI] [PubMed] [Google Scholar]
- Moll H, Fuchs H, Blank C, Rollinghoff M. Langerhans cells transport Leishmania major from the infected skin to the draining lymph node for presentation to antigen-specific T cells. Eur J Immunol. 1993;23:1595–1601. doi: 10.1002/eji.1830230730. [DOI] [PubMed] [Google Scholar]
- Murphy DB, Rath S, Pizzo E, Rudensky AY, George A, Larson JK, Janeway CA., Jr Monoclonal antibody detection of a major self peptide. MHC class II complex. J Immunol. 1992;148:3483–3491. [PubMed] [Google Scholar]
- Nakano H, Lin KL, Yanagita M, Charbonneau C, Cook DN, Kakiuchi T, Gunn MD. Blood-derived inflammatory dendritic cells in lymph nodes stimulate acute T helper type 1 immune responses. Nat Immunol. 2009;10:394–402. doi: 10.1038/ni.1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Netea MG, Brown GD, Kullberg BJ, Gow NA. An integrated model of the recognition of Candida albicans by the innate immune system. Nat Rev Microbiol. 2008;6:67–8. doi: 10.1038/nrmicro1815. [DOI] [PubMed] [Google Scholar]
- Ohl L, Mohaupt M, Czeloth N, Hintzen G, Kiafard Z, Zwirner J, Blankenstein T, Henning G, Forster R. CCR7 governs skin dendritic cell migration under inflammatory and steady-state conditions. Immunity. 2004;21:279–288. doi: 10.1016/j.immuni.2004.06.014. [DOI] [PubMed] [Google Scholar]
- Probst HC, Tschannen K, Odermatt B, Schwendener R, Zinkernagel RM, Van Den Broek M. Histological analysis of CD11c-DTR/GFP mice after in vivo depletion of dendritic cells. Clin Exp Immunol. 2005;141:398–404. doi: 10.1111/j.1365-2249.2005.02868.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romani L. Cell mediated immunity to fungi: a reassessment. Med Mycol. 2008;46:515–529. doi: 10.1080/13693780801971450. [DOI] [PubMed] [Google Scholar]
- Rudensky A, Rath S, Preston-Hurlburt P, Murphy DB, Janeway CA., Jr On the complexity of self. Nature. 1991;353:660–662. doi: 10.1038/353660a0. [DOI] [PubMed] [Google Scholar]
- Segura E, Villadangos JA. Antigen presentation by dendritic cells in vivo. Curr Opin Immunol. 2009;21:105–110. doi: 10.1016/j.coi.2009.03.011. [DOI] [PubMed] [Google Scholar]
- Serbina NV, Jia T, Hohl TM, Pamer EG. Monocyte-mediated defense against microbial pathogens. Annu Rev Immunol. 2008;26:421–452. doi: 10.1146/annurev.immunol.26.021607.090326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinman RM. Dendritic cells in vivo: a key target for a new vaccine science. Immunity. 2008;29:319–324. doi: 10.1016/j.immuni.2008.08.001. [DOI] [PubMed] [Google Scholar]
- Villadangos JA, Schnorrer P. Intrinsic and cooperative antigen-presenting functions of dendritic-cell subsets in vivo. Nat Rev Immunol. 2007;7:543–555. doi: 10.1038/nri2103. [DOI] [PubMed] [Google Scholar]
- Wüthrich M, Filutowicz HI, Klein BS. Mutation of the WI-1 gene yields an attenuated Blastomyces dermatitidis strain that induces host resistance. J Clin Invest. 2000;106:1381–1389. doi: 10.1172/JCI11037. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.