Abstract
Background
Opioidergic neurotransmission is critical in many, and possibly all, forms of substance dependence, and several opioid-system genes have been shown previously to be associated with substance dependence disorders. The pro-opiomelanocortin gene (POMC) encodes several peptides important for endogenous opioidergic neurotransmission. The authors tested whether POMC genetic variation affects risk for substance dependence.
Methods
Five single nucleotide polymorphisms spanning POMC were examined in independent family and case-control samples. Family-based studies included 854 subjects from 319 African American (AA) families and 761 subjects from 313 European American (EA) families. Each family had a pair of siblings affected with cocaine and/or opioid dependence. Case-control studies included 791 cases (455 AAs and 336 EAs) affected with alcohol, cocaine, and/or opioid dependence and 682 controls (199 AAs and 483 EAs).
Results
Family-based analyses revealed an association of rs6719226 with opioid dependence in AA families, and rs6713532 with cocaine dependence in EA families (P=0.010–0.044). Case-control analyses demonstrated an association of rs6713532 with alcohol or cocaine dependence in EAs (Pallele-wise=0.003–0.008). Moreover, the minor allele of rs1866146 was found to be a risk factor for cocaine or opioid dependence in AAs (Pallele-wise=0.010–0.017), and for alcohol, cocaine or opioid dependence in EAs (Pallele-wise=0.001–0.003). Logistic regression analyses in which sex and age were considered, and population stratification analyses, confirmed these findings. Additionally, specific haplotypes increased risk for cocaine dependence (P=0.023) in AAs, or opioid dependence (P=0.012) in EAs.
Conclusions
Based on these replicated results, we conclude that variation in POMC confers vulnerability to multiple forms of substance dependence.
Keywords: Pro-opioimelanocortin gene, alcohol or drug dependence, family-based study, case-control study, logistic regression, population stratification
Introduction
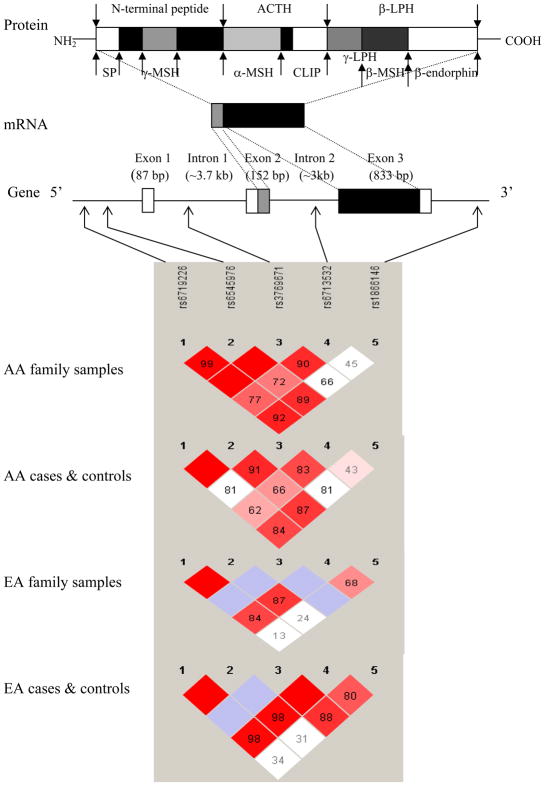
Pro-opiomelanocortin (POMC), encoded by the polycistronic POMC gene (or POMC) on chromosome 2p23.3, is a polypeptide precursor protein with 241 amino acid residues. As many as 10 functionally different peptides can be derived from POMC via extensive tissue-specific post-translational processing. These peptides include adrenocorticotropin (ACTH), β- and γ-lipotropin (β- and γ-LPH), α-, β-, and γ-melanocyte-stimulating hormone (α-, β-, and γ-MSH), corticotropin-like intermediate lobe peptide (CLIP), and β-endorphin (Figure 1). These peptides play crucial roles in numerous biological processes such as pain (1), energy homeostasis (2), melanocyte stimulation (3), and immune modulation (4).
Figure 1.
POMC, coded peptides, and marker pair-wise linkage disequilibrium (LD) plots in African Americans (AAs) and European Americans (EAs).
POMC, pro-opiomelanocortin; SP, signal peptide; ACTH: adrenocorticotropin; β- and γ-LPH, β- and γ-lipotropin; α-, β- and γ-MSH, α-, β- and γ-melanocyte-stimulating hormone; CLIP, corticotropin like intermediate lobe peptide. LD between pairs of markers was measured by D′ values (0 ~ 1), as shown in the squares.
Among these biologically active peptides, ACTH and β-endorphin are two principal components of the hypothalamic-pituitary-adrenal (HPA) axis. ACTH mediates the stress response in vertebrates (5). Stress induces the secretion of corticotropin-release hormone (CRH) from the hypothalamus, which stimulates ACTH synthesis and release from the anterior pituitary. ACTH, in turn, promotes the release of glucocorticoids (e.g, cortisol, a major stress hormone) from the adrenal cortex. Through negative feedback, glucocorticoids regulate the expression of CRH and ACTH. β-endorphin, an endogenous opioid peptide exerting potent analgesic and euphoric effects through interaction with opioid receptors, is made in neurons of the brain stem, as well as those in the hypothalamus and pituitary. It produces behavioral effects similar to exogenous opioids and is released in the nucleus accumbens (NAc), the major brain reward center (6). Based on these actions, both ACTH and β-endorphin have been implicated in craving use of drugs and alcohol (7).
There is consistent evidence that stress, HPA function, and addictive behaviors are closely interrelated. Studies in rodents have shown that physiological or psychological stressors elevate POMC mRNA levels in the pituitary (8), and chronic antidepressant treatment decrease pituitary levels of POMC mRNA (9). Stress is a common trigger for drug or alcohol use, and individuals experiencing stress (or depression and/or anxiety) are more prone to abuse drugs or alcohol. Consumption of drugs or alcohol can, in turn, alter HPA function, and modulate the production of ACTH, β-endorphin, or glucocorticoid hormones. Cocaine administration activates the opioid system and the HPA axis, and enhances the secretion of β-endorphin, ACTH and corticosterone in rat (10), mouse (11), and human (12). Similarly, acute ethanol administration induces an increase in the release of pituitary β-endorphin (13). In addition, both increased and decreased β-endorphin release has been observed in chronic ethanol treatment (14).
POMC-derived peptides also actively regulate alcohol- and drug-related behaviors. A significant inverse correlation was observed between baseline cortisol levels and craving for alcohol in alcohol-dependent subjects, and treatment with naltrexone (an antagonist at μ-, δ- and κ-opioid receptors) increased plasma cortisol and ACTH levels and decreased alcohol craving and consumption (15). Moreover, enhanced ACTH and cortisol may contribute to alcohol withdrawal symptoms such as stress and depression. Plasma levels of these hormones were elevated immediately after alcohol withdrawal, but normalized over the following two weeks as the severity of withdrawal symptoms decreased (16). In addition, lower cortisol levels were associated with an increased risk of alcoholic relapse (17). Plasma levels of β-endorphin were also reduced during alcohol withdrawal but remained low even when withdrawal symptoms subsided (18). This implies that the reduced level of β-endorphin in alcoholics is unlikely due to alcohol withdrawal, but may be a trait contributing to alcohol craving and consumption. Therefore, a baseline deficiency of β-endorphin is of potential importance in the initiation and maintenance of drug-taking behaviors (19).
Given the diverse function of POMC-derived peptides, POMC variation may affect the risk for several disease states. Peptides such as α-MSH, ACTH, and lipotrophin are involved in appetite regulation, glucose and lipid metabolism, and energy homeostasis. Polymorphisms at POMC may reasonably contribute to the development of obesity. Studies have shown an association between four POMC polymorphisms (rs1009388, rs2071345, rs1042571, and rs1866146) and obesity traits in the general population (20). In addition, α-MSH has a dual role in regulating food intake and influencing hair pigmentation, and ACTH stimulates the release of glucocorticoids and maintains adrenal activity. Rare loss-of-function mutations in POMC were found to cause severe early-onset obesity, adrenal insufficiency, and red hair pigmentation (21). The reinforcing effect of β-endorphin and its release from the hypothalamus or the pituitary to the nucleus accumbens (NAc) suggest a general involvement of β-endorphin in substance use disorders.
Numerous studies have suggested, and twin studies (22) established, that the genetic contributions to risk for substance dependence are both shared (i.e., common to multiple substances) and specific (i.e., applicable to only one substance). POMC, which encodes several peptides (including ACTH and β-endorphin), can be viewed as, potentially, a common susceptibility gene for substance dependence disorders. To date, one family-based study (23) has demonstrated an association between POMC variants (two SNPs in intron 1) and opioid dependence, and one case-control-based study (24) (by haplotype analyses) has provided evidence for a sex-specific role of POMC in ethanol preference and dependence.
To examine the potential role of POMC variation in the risk for drug or alcohol dependence, we first conducted a family-based study in which we analyzed the association of five POMC single nucleotide polymorphisms (SNPs) with drug and/or alcohol dependence in both African Americans (AAs) and European Americans (EAs). We then replicated the results of that study using independent AA and EA case-control samples.
Methods and Materials
Sample Description
Four sets of samples (both AA and EA family and case-control samples) were collected (Table 1). Subjects were recruited at four sites in the US: University of Connecticut Health Center (Farmington, CT, USA), Yale University School of Medicine (APT Foundation, New Haven, CT, USA), Medical University of South Carolina (Charleston, SC, USA), and McLean Hospital (Harvard Medical School, Belmont, MA, USA). The study protocol was approved by the institutional review board (IRB) at each clinical site. After complete description of the study to the subjects, written informed consent was obtained.
Table 1.
Characteristics of African American and European American Family and Case-control Samples
| Characteristic | African American (AA) | European American (EA) | ||
|---|---|---|---|---|
| Family sample: | ||||
| Number of families | 319 | 313 | ||
| Number of parents | 70 | 89 | ||
| Number of all family members | 854a | 761a | ||
| Mean | SD | Mean | SD | |
| Age (years) | 40.9 | 7.3 | 37.8 | 10.2 |
| N | % | N | % | |
| Sex (% female) | 504 | 59.0 | 366 | 48.1 |
| Number (%) of subjects with cocaine dependencec | 676 | 79.2 | 559 | 73.5 |
| Number (%) of subjects with opioid dependencec | 214 | 25.1 | 252 | 33.1 |
| Number (%) of subjects with alcohol dependencec | 328 | 38.4 | 323 | 42.5 |
| Case-control sample: | ||||
| Controls | ||||
| Number of subjects | 199 (151a + 48b) | 483 (126a + 357b) | ||
| Mean | SD | Mean | SD | |
| Age (years) | 36.1 | 13.3 | 30.0 | 11.2 |
| N | % | N | % | |
| Sex (% female) | 151 | 75.9 | 285 | 59.0 |
| Cases | ||||
| Number of subjects | 455a | 336a | ||
| Mean | SD | Mean | SD | |
| Age (years) | 40.9 | 9.3 | 40.3 | 11.8 |
| N | % | N | % | |
| Sex (% female) | 180 | 39.6 | 123 | 36.6 |
| Number (%) of subjects with cocaine dependencec | 305 | 67.2 | 195 | 58.0 |
| Number (%) of subjects with opioid dependencec | 80 | 17.6 | 144 | 42.8 |
| Number (%) of subjects with alcohol dependencec | 379 | 83.5 | 278 | 82.7 |
All family members, 151 AA controls, 455 AA cases, 126 EA controls, and 336 EA cases were evaluated with the Semi-Structured Assessment for Drug Dependence and Alcoholism (SSADDA) instrument.
48 AA controls and 357 EA controls were evaluated with the Structural Clinical Interview for DSM-III-R (SCID-III-R) or DSM-IV (SCID-IV).
Besides affected with the indicated disorder, some subjects had co-morbid other substance dependence disorders
Information about the two sets of family samples was provided previously (25). Briefly, 854 subjects from 319 AA families and 761 subjects from 313 EA families were recruited. Each family included a sibling pair where both siblings were affected (at a minimum) with cocaine and/or opioid dependence (CD and/or OD). Subjects were interviewed using a computer-assisted version of the Semi-structured Assessment for Drug Dependence and Alcoholism (SSADDA) instrument (26). Individuals with a primary diagnosis of a major psychotic illness (e.g., schizophrenia or schizoaffective disorder) were excluded as probands. Additional siblings and parents of affected sibling pairs (ASPs) were recruited when possible regardless of their affection status. Family subjects were classified as genetically AAs or EAs based on a Bayesian model-based clustering method using the short tandem repeat (STR) marker linkage set as previously described (27).
Two sets of case-control samples (AA: 199 controls and 455 cases; EA: 483 controls and 336 cases) were recruited. All AA and EA cases, 151 AA controls, and 126 EA controls were assessed with the SSADDA instrument (26). 48 AA controls and 357 EA controls were evaluated using the Structured Clinical Interview for DSM-III-R (SCID-III-R) (American Psychiatric Association, 1987) or DSM-IV (SCID-IV) (American Psychiatric Association, 1994), as described previously (28). Cases were diagnosed with alcohol dependence (AD), CD, and/or OD using the SSADDA instrument. Control subjects were screened to exclude those with major Axis I mental disorders, including alcohol or drug dependence, mood disorders, major anxiety disorders, and psychotic disorders. In general, the control subjects were younger and more likely to be female than the case subjects. The ethnicity of case and control subjects was self-identified. The genetic background of a majority of case and control subjects was also analyzed using a set of ancestry-informative markers (AIMs) (see below).
Marker Selection and Genotyping
Five SNPs, which are evenly distributed in the POMC region (average inter-marker distance: 3,860 bp), were selected from the NCBI dbSNP database (http://www.ncbi.nlm.nih.gov/SNP) (Table 2). Of these SNPs, two (rs6719226 and rs6545976) are in the upstream region, one (rs3769671) is in intron 1, one (rs6713532) is in intron 2, and one (rs1866146) is in the downstream region. As an additional measure of SNP coverage, we downloaded the HapMap CEU and YRI genotype data of SNPs located in the POMC gene region (chr2:25234077–25249516) (only nine SNPs, including the five SNPs we studied, with minor allele frequency > 2%), and then we evaluated whether these five SNPs can capture most information from the other four SNPs using the Tagger program in Haploview 4.0 (29). These five SNPs gave a mean r2 of 0.70 with the nine HapMap SNPs in the CEU (CEPH Europeans) population and 0.85 with the nine HapMap SNPs in the YRI (Africans) population. Thus, these five SNPs can potentially capture most of the genetic information of all other markers in the POMC region.
Table 2.
Characteristics of SNPs in the POMC Region on Chromosome 2
| POMC Markers | SNP Location | Chromosome positiona (bp) | Intermarker Distance (bp) | Allele | MAFb HapMap-YRI | MAFb HapMap-CEU |
|---|---|---|---|---|---|---|
| rs6719226 | 5′ flanking region | 25249516 | – | C/G | 0.38 (G) | 0.06 (G) |
| rs6545976 | 5′ flanking region | 25248154 | 1362 | G/T | 0.39 (G) | 0.06 (T) |
| rs3769671 | Intron 1 | 25243657 | 4497 | A/C | 0.03 (C) | 0.05 (C) |
| rs6713532 | Intron 2 | 25238337 | 5320 | C/T | 0.37 (T) | 0.22 (C) |
| rs1866146 | 3′ flanking region | 25234077 | 4260 | C/T | 0.09 (C) | 0.29 (C) |
Chromosome positions are based on Homo sapiens chromosome 2 genomic contig NT_022184.14.
MAF, minor allele frequency in the HapMap database; YRI, Yoruba in Ibadan, Nigeria; CEU, CEPH Utah residents with ancestry from northern and western Europe.
DNA was extracted from immortalized cell lines or directly from fresh blood or saliva. All markers [except rs1866146, which was genotyped by the TaqMan technique (30)] were genotyped by the Illumina GoldenGate Assay methodology (Illumina Inc., San Diego, CA, USA). To identify the genetic background of the unrelated subjects, we genotyped an additional 38 AIMs, including 37 short tandem repeat markers (STRs) and SNP marker FY (31), in a majority of the unrelated subjects (139 of 151 AA controls, 444 of 455 AA cases, 112 of 126 EA controls, and 316 of 336 EA cases) which were assessed with the SSADDA instrument. Genotype data for the 38 AIMs were available for all SCID-III-R or SCID-IV assessed control subjects (48 AAs and 357 EAs) (28,31). In total, there were AIM data for 96.1% (1,416/1,473) of the unrelated subjects.
Statistical Analysis
For family samples, Mendelian inheritance of all genotypes was evaluated using PedCheck (32). Genotype distribution deviation from Hardy-Weinberg equilibrium (HWE) and pair-wise marker linkage disequilibrium (LD) were examined using the program Haploview 4.0 (29), with genotype data from parents. Marker allele transmission from heterozygous parents to affected offspring (or marker allele sharing between affected sibling pairs) was examined using the Family-based Association Test (FBAT) program (33), assuming an additive model. Haplotype frequencies were estimated in the founders (or parents) and haplotype association analyses were performed using the Haplotype-based Association Test (HBAT) (34).
For case-control samples, the ancestry proportion of each subject was estimated using the genotype data of 38 AIMs through application of the program STRUCTURE (35,36). Pair-wise marker LD analyses were performed using the program Haploview 4.0 (29) in cases and controls separately and combined. HWE analyses, allele and genotype frequency calculation, and individual marker association analysis were performed using the program PowerMarker (37). Estimation of haplotype frequencies and examination of the effect of haplotypes on disease traits were conducted using haplotype trend regression (HTR) implemented in HelixTree software (Golden Helix, Inc., Bozeman, MT, USA). Potential confounding effects of sex and age (not matched in our cases and controls) on individual marker or haplotype association results were analyzed using a backward stepwise logistic regression (LR) analysis implemented in SPSS 15.0 (SPSS Inc, Chicago, IL, USA), as described previously (28). The influence of POMC variants on the three disease traits (AD, CD, or OD) was investigated under three models (additive, dominant, and recessive). The disease trait was the dependent variable, and sex, age, and the probabilities of alleles, genotypes, or haplotypes were served as covariates. The probability of each haplotype for each subject was approximated using the logistic regression analysis incorporated in the HelixTree software.
Results
HWE, Marker Pair-wise LD, Population Stratification Analysis Results
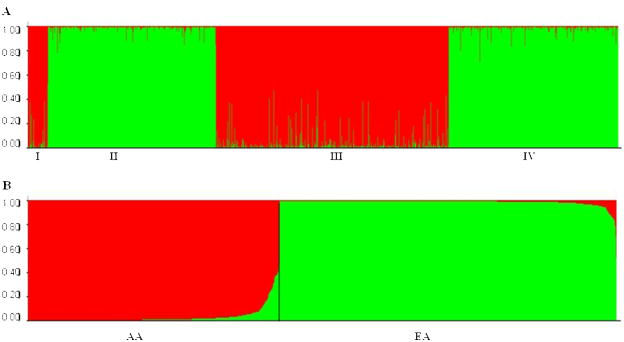
Genotype distributions at all polymorphisms were consistent with HWE expectations in genotyped founders (or parents) in both AA and EA families, and in genotyped unrelated AA or EA cases or controls, except the genotype distribution of rs6713532 in EA controls [P=0.039; non-significant when corrected for multiple comparisons at α level of 0.010 (α=0.050/5)]. Substantial pair-wise LD was observed between adjacent markers (rs6719226 to rs6713532) in both AA and EA samples (Figure 1). A moderate LD was observed between rs6713532 and rs1866146 in EA samples. Ancestry proportions of four groups of unrelated subjects are shown in Figure 2. Genetically identified ethnicity was consistent with self-reported ethnicity (African ancestry proportions >50% for all AA subjects, and European ancestry proportions >50% for all EA subjects). The degree of admixture of the European ancestry (the green part) in AA subjects was only 3.3%, and the degree of admixture of the African ancestry (the red part) in EA subjects was only 1.3%. The low estimated admixture rates in AAs and EAs probably reflect in part the statistical effects of the lack of a non-admixed African sample in the cluster analysis.
Figure 2.
Ancestry structure of African Americans (AAs) and European Americans (EAs). (A) Ancestry proportions of four groups of subjects (Group I = 48 SCID-III-R or SCID-IV evaluated AA controls, Group II = 357 SCID-III-R or SCID-IV evaluated EA controls, Group III = 444 SSADDA assessed AA cases and 139 SSADDA assessed AA controls, and Group IV = 316 SSADDA assessed EA cases and 112 SSADDA assessed EA controls). (B) Subjects are sorted by African ancestry proportions (red) and European ancestry proportions (green). X axis = individuals; Y axis = ancestry proportions.
Family-based Association Analysis Results
In AA families, rs6719226 major allele (C) was transmitted significantly more frequently to offspring with OD than expected (P=0.010, which could withstand the Bonferroni correction at the level of α=0.050/5=0.010). In EA families, rs6713532 minor allele (C) was transmitted more often to offspring with CD than by chance, and a marginally significant association was found between this marker and CD (P=0.044) (Table 3). To examine further the evidence for association of POMC with AD, CD, or OD, haplotype-based analyses including all five markers were performed. However, none of these haplotypes was found to be over transmitted to affected offspring (Supplementary Material Table S1).
Table 3.
Family-based Association Tests (additive model) with SNPs in the POMC Region
| POMC Markers | Allele Freq (in AAs) | AA-AD P-allelea | AA-CD P-allelea | AA-OD P-allelea | Allele Freq (in EAs) | EA-AD P-allelea | EA-CD P-allelea | EA-OD P-allelea |
|---|---|---|---|---|---|---|---|---|
| rs6719226 | 0.34 (G) | 0.469 (53) | 0.772 (32) | 0.010b (21) | 0.05 (G) | 0.823 (9) | 0.263 (9) | 0.401 (11) |
| rs6545976 | 0.45 (T) | 0.366 (57) | 0.493 (41) | 0.125 (24) | 0.06 (T) | 0.419 (11) | 0.264 (9) | 0.590 (12) |
| rs3769671 | 0.06 (C) | 0.293 (17) | 0.078 (12) | 0.238 (8) | 0.05 (C) | 0.756 (9) | 0.460 (5) | 1.000 (7) |
| rs6713532 | 0.49 (T) | 0.603 (56) | 0.080 (45) | 0.244 (31) | 0.27 (C) | 0.550 (37) | 0.044 (24) | 0.713 (35) |
| rs1866146 | 0.14 (C) | 0.149 (33) | 0.184 (29) | 0.932 (15) | 0.34 (C) | 0.896 (44) | 0.433 (34) | 1.000 (37) |
Abbreviations: AA (or EA)-AD, AA (or EA) families with siblings affected with AD; AA (or EA)-CD, AA (or EA) families with siblings affected with CD; AA (or EA)-OD, AA (or EA) families with siblings affected with OD.
P values for allele transmission (numbers in parentheses are numbers of informative families).
The P value remained significant after the Bonferroni correction at the level of α=0.05/5=0.010.
Note: statistically significant P-values are in bold.
Case-control Association Analysis Results
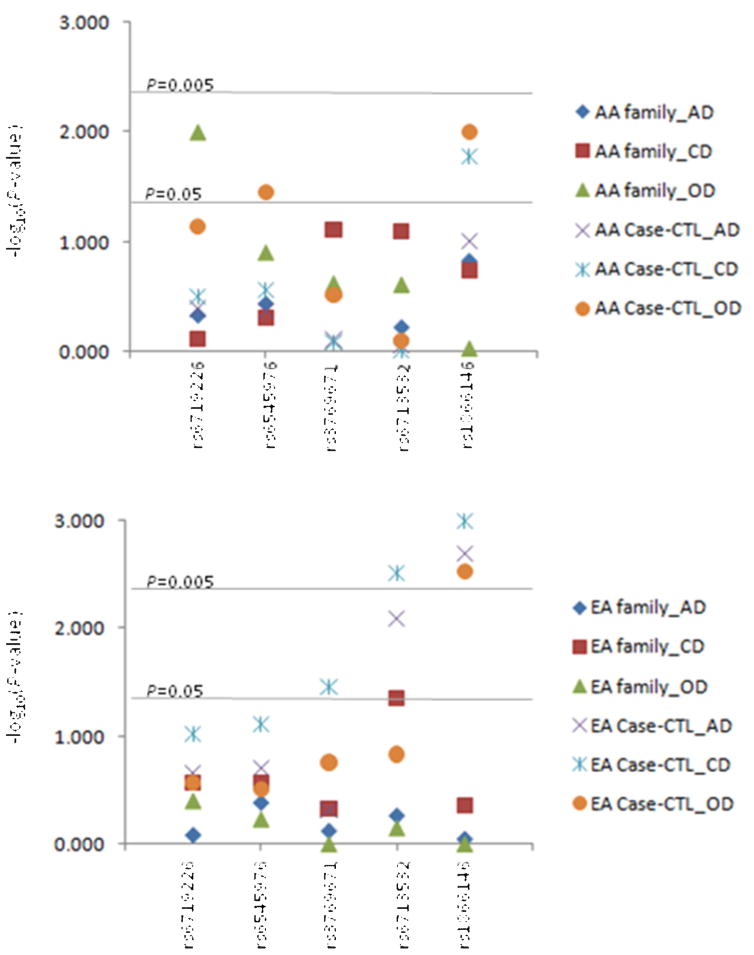
Individual marker case-control analysis results are given in Table 4. In AA samples, the frequency of rs6719226 major allele (C) was non-significantly more common in OD cases (72.0%) than in controls (65.0%) (Pallele-wise=0.072). Rs6545976 major allele (G) was significantly more common in OD cases (61.0%) than in controls (51.0%) (Pallele-wise=0.035). The frequency of rs1866146 minor allele (C) was significantly higher in CD and OD cases (13.0% and 16.0%, respectively) as compared to controls (8.0%) (CD vs. Con: Pallele-wise=0.017; OD vs. Con: Pallele-wise=0.010). In EA samples, a nominally-significant association was noted between rs3769671 and CD (Pallele-wise=0.035). Rs6713532 minor allele (C) was significantly more frequent in AD and OD cases (26.0% and 27.0%, respectively) as compared to controls (20.0%) (AD vs. Con: Pallele-wise=0.008; CD vs. Con: Pallele-wise=0.003) (both associations withstood the Bonferroni correction at the level of α=0.050/5=0.010). Similarly, there was an increased frequency of rs1866146 minor allele (C) in AD, CD, and OD cases (35.0%, 39.0%, and 37.0%, respectively) than in controls (28.0%) (AD vs. Con: Pallele-wise=0.002; CD vs. Con: Pallele- wise=0.001; OD vs. Con: Pallele-wise=0.003). The positive association between rs1866146 and OD in AAs or all three phenotypes in EAs survived a Bonferroni correction (α=0.050/5=0.010). Figure 3 summarizes the association significance (denoted as −log10P-value) of five POMC SNPs in AD, CD, and OD from both family- and case-control-based studies using samples collected from the two populations (AAs and EAs).
Table 4.
POMC Marker Allele/Genotype Frequency Comparison between Cases and Controls
| POMC Markers | AA-Con | AA-AD | Pa | AA-CD | Pa | AA-OD | Pa | EA-Con | EA-AD | Pa | EA-CD | Pa | EA-OD | Pa |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| n=199 | n=379 | n=305 | n=80 | n=483 | n=278 | n=195 | n=144 | |||||||
| Freq | Freq | Freq | Freq | Freq | Freq | Freq | Freq | |||||||
| rs6719226 | ||||||||||||||
| C | 0.65 | 0.67 | 0.412 | 0.68 | 0.320 | 0.72 | 0.072 | 0.96 | 0.95 | 0.223 | 0.94 | 0.095 | 0.94 | 0.272 |
| G | 0.35 | 0.33 | 0.32 | 0.28 | 0.04 | 0.05 | 0.06 | 0.06 | ||||||
| C/C | 0.43 | 0.45 | 0.640 | 0.47 | 0.614 | 0.55 | 0.195 | 0.92 | 0.90 | 0.269 | 0.88 | 0.119 | 0.90 | 0.152 |
| C/G | 0.43 | 0.43 | 0.42 | 0.35 | 0.08 | 0.10 | 0.11 | 0.10 | ||||||
| G/G | 0.14 | 0.11 | 0.12 | 0.10 | 0.00 | 0.00 | 0.01 | 0.00 | ||||||
| rs6545976 | ||||||||||||||
| G | 0.51 | 0.53 | 0.438 | 0.54 | 0.278 | 0.61 | 0.035 | 0.96 | 0.94 | 0.201 | 0.94 | 0.077 | 0.94 | 0.308 |
| T | 0.49 | 0.47 | 0.46 | 0.39 | 0.04 | 0.06 | 0.06 | 0.06 | ||||||
| G/G | 0.29 | 0.27 | 0.111 | 0.29 | 0.111 | 0.36 | 0.083 | 0.92 | 0.89 | 0.445 | 0.88 | 0.160 | 0.89 | 0.508 |
| G/T | 0.44 | 0.52 | 0.51 | 0.49 | 0.08 | 0.11 | 0.11 | 0.10 | ||||||
| T/T | 0.27 | 0.21 | 0.20 | 0.15 | 0.00 | 0.00 | 0.01 | 0.01 | ||||||
| rs3769671 | ||||||||||||||
| A | 0.96 | 0.96 | 0.792 | 0.96 | 0.835 | 0.98 | 0.307 | 0.96 | 0.97 | 0.493 | 0.98 | 0.035 | 0.98 | 0.174 |
| C | 0.04 | 0.04 | 0.04 | 0.02 | 0.04 | 0.03 | 0.02 | 0.02 | ||||||
| A/A | 0.93 | 0.93 | 0.796 | 0.93 | 0.890 | 0.96 | 0.606 | 0.93 | 0.94 | 0.485 | 0.97 | 0.032 | 0.96 | 0.166 |
| A/C | 0.06 | 0.07 | 0.07 | 0.04 | 0.07 | 0.06 | 0.03 | 0.04 | ||||||
| C/C | 0.01 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | ||||||
| rs6713532 | ||||||||||||||
| C | 0.51 | 0.52 | 0.884 | 0.51 | 0.987 | 0.50 | 0.787 | 0.20 | 0.26 | 0.008b | 0.27 | 0.003 b | 0.24 | 0.147 |
| T | 0.49 | 0.48 | 0.49 | 0.50 | 0.80 | 0.74 | 0.73 | 0.76 | ||||||
| C/C | 0.27 | 0.28 | 0.981 | 0.27 | 0.950 | 0.28 | 0.844 | 0.05 | 0.07 | 0.015 | 0.08 | 0.011 | 0.07 | 0.349 |
| C/T | 0.48 | 0.48 | 0.49 | 0.44 | 0.29 | 0.38 | 0.38 | 0.34 | ||||||
| T/T | 0.25 | 0.24 | 0.24 | 0.28 | 0.66 | 0.55 | 0.54 | 0.59 | ||||||
| rs1866146 | ||||||||||||||
| C | 0.08 | 0.11 | 0.100 | 0.13 | 0.017 | 0.16 | 0.010 b | 0.28 | 0.35 | 0.002 b | 0.39 | 0.001 b | 0.37 | 0.003 b |
| T | 0.92 | 0.89 | 0.87 | 0.84 | 0.72 | 0.65 | 0.61 | 0.63 | ||||||
| C/C | 0.01 | 0.01 | 0.253 | 0.01 | 0.055 | 0.01 | 0.029 | 0.10 | 0.11 | 0.002 b | 0.15 | 0.001 b | 0.15 | 0.017 |
| C/T | 0.15 | 0.21 | 0.24 | 0.29 | 0.36 | 0.48 | 0.47 | 0.44 | ||||||
| T/T | 0.84 | 0.78 | 0.75 | 0.70 | 0.54 | 0.41 | 0.38 | 0.41 | ||||||
Abbreviations: AA (or EA)-Con, AA (or EA) controls; AA (or EA)-AD, AA (or EA) cases with alcohol dependence (AD); AA (or EA)-CD, AA (or EA) cases with cocaine dependence (CD); AA (or EA)-OD, AA (or EA) cases with opioid dependence (OD).
P values obtained by comparing allele or genotype frequencies between cases and controls;
P values remained significant after the Bonferroni correction at the level of α=0.05/5=0.010.
Note: statistically significant P-values are in bold.
Figure 3.
Plot of association significance (denoted as −log10P-value) of five POMC SNPs in AD, CD and OD from both family- and case-control-based studies.
AA: African American; EA: European American; family: family-based association test (FBAT); Case-CTL: case-control-based association analysis (allele-wise); AD: alcohol dependence; CD: cocaine dependence; OD: opioid dependence.
No significant difference in haplotype frequency distributions was detected between AA cases and controls. However, the most common haplotype C-G-A-T-T, which was comprised of major alleles of all five POMC SNPs, was significantly less frequent in EA cases affected with CD (56.0%) than in EA controls (65.0%) (P-haplo=0.028) (Supplementary Material Table S2).
Results of backward stepwise logistic regression (LR) analyses are summarized in Table 5. In AA samples, rs1866146 minor allele (C) showed a trend for an effect on risk of CD under either an additive model (P=0.054) or a dominant model (P=0.054). Haplotype C-G-A-C-C, the only haplotype (with a frequency of at least 5%) harboring rs1866146 minor allele (C) in AAs (see Supplementary Material Table S2), increased the risk for CD (P=0.023). In EA samples, rs1866146 minor allele (C) increased the risk for AD, CD, or OD, irrespective of whether an additive or a recessive model was considered [additive model: P=0.003 (AD), P<0.001 (CD), and P=0.003 (OD); dominant model: P=0.001 (AD), P<0.001 (CD), and P=0.002 (OD)]. The most common haplotype, C-G-A-T-T, consisting of major alleles of all five POMC SNPs, reduced the risk for both AD (P=0.007) and CD (P=0.001). Additionally, the third most frequent haplotype, C-G-A-T-C [harboring rs1866146 minor allele (C), see Supplementary Material Table S2], increased the risk for OD (P=0.012).
Table 5.
Backward Stepwise Logistic Regression (LR) Analysis of POMC SNPs in Cases and Controls
| Variables | AA-AD vs. AA-Con | AA-CD vs. AA-Con | AA-OD vs. AA-Con | ||||||
|---|---|---|---|---|---|---|---|---|---|
| P | β | OR | P | β | OR | P | β | OR | |
| Genotype LR (additive) | |||||||||
| Male | < 0.001 | 1.60 | 4.98 | < 0.001 | 1.44 | 4.22 | < 0.001 | 2.08 | 8.02 |
| Age | < 0.001 | 0.04 | 1.04 | < 0.001 | 0.06 | 1.06 | < 0.001 | 0.06 | 1.07 |
| rs3769671^C | - | - | - | - | - | - | 0.095 | −2.19 | 0.11 |
| rs1866146^C | - | - | - | 0.054 | 0.94 | 2.57 | - | - | - |
| Genotype LR (dominant) | |||||||||
| Male | < 0.001 | 1.60 | 4.98 | < 0.001 | 1.44 | 4.23 | < 0.001 | 2.00 | 7.40 |
| Age | < 0.001 | 0.04 | 1.04 | < 0.001 | 0.06 | 1.06 | < 0.001 | 0.07 | 1.07 |
| rs6719226^G | - | - | - | - | - | - | 0.066 | −0.59 | 0.56 |
| rs1866146^C | - | - | - | 0.054 | 0.50 | 1.65 | - | - | - |
| Genotype LR (recessive) | |||||||||
| Male | < 0.001 | 1.60 | 4.98 | < 0.001 | 1.45 | 4.27 | < 0.001 | 2.02 | 7.52 |
| Age | < 0.001 | 0.04 | 1.04 | < 0.001 | 0.06 | 1.06 | < 0.001 | 0.06 | 1.07 |
| Haplotype LR | |||||||||
| Male | < 0.001 | 1.60 | 4.97 | < 0.001 | 1.46 | 4.31 | < 0.001 | 2.05 | 7.75 |
| Age | < 0.001 | 0.04 | 1.04 | < 0.001 | 0.06 | 1.06 | < 0.001 | 0.06 | 1.07 |
| C-G-A-C-C | - | - | - | 0.023 | 1.40 | 4.06 | - | - | - |
| Variables | EA-AD vs. EA-Con | EA-CD vs. EA-Con | EA-OD vs. EA-Con | ||||||
|---|---|---|---|---|---|---|---|---|---|
| P | β | OR | P | β | OR | P | β | OR | |
| Genotype LR (additive) | |||||||||
| Male | < 0.001 | 0.93 | 2.53 | < 0.001 | 0.80 | 2.22 | < 0.001 | 0.90 | 2.45 |
| Age | < 0.001 | 0.07 | 1.07 | < 0.001 | 0.07 | 1.07 | < 0.001 | 0.05 | 1.06 |
| rs3769671^C | - | - | - | 0.093 | −1.68 | 0.18 | - | - | - |
| rs1866146^C | 0.003 | 0.79 | 2.20 | < 0.001 | 1.01 | 2.75 | 0.003 | 0.62 | 1.87 |
| Genotype LR (dominant) | |||||||||
| Male | < 0.001 | 0.93 | 2.52 | < 0.001 | 0.81 | 2.27 | < 0.001 | 0.89 | 2.43 |
| Age | < 0.001 | 0.07 | 1.07 | < 0.001 | 0.07 | 1.07 | < 0.001 | 0.05 | 1.05 |
| rs3769671^C | - | - | - | 0.093 | −0.84 | 0.43 | - | - | - |
| rs1866146^C | 0.001 | 0.62 | 1.85 | < 0.001 | 0.74 | 2.01 | 0.002 | 0.92 | 2.52 |
| Genotype LR (recessive) | |||||||||
| Male | < 0.001 | 0.91 | 2.49 | < 0.001 | 0.78 | 2.18 | < 0.001 | 0.86 | 2.35 |
| Age | < 0.001 | 0.07 | 1.07 | < 0.001 | 0.07 | 1.07 | < 0.001 | 0.05 | 1.05 |
| rs1866146^C | - | - | - | 0.055 | 0.56 | 1.75 | 0.059 | 0.59 | 1.80 |
| Haplotype LR | |||||||||
| Male | < 0.001 | 0.98 | 2.66 | < 0.001 | 0.85 | 2.35 | < 0.001 | 0.95 | 2.60 |
| Age | < 0.001 | 0.08 | 1.08 | < 0.001 | 0.07 | 1.08 | < 0.001 | 0.06 | 1.06 |
| C-G-A-T-T | 0.007 | −0.68 | 0.50 | 0.001 | −0.93 | 0.40 | - | - | - |
| C-G-A-C-C | - | - | - | - | - | - | 0.079 | 0.66 | 1.93 |
| C-G-A-T-C | - | - | - | - | - | - | 0.012 | 1.13 | 3.09 |
Abbreviations: AA (or EA)-Con, AA (or EA)-AD, AA (or EA)-CD, and AA (or EA)-OD are the same as in Table 3; β, regression coefficient; OR, odds ratio.
Note: only the covariates that were retained in the final regression step are listed, and statistically significant P-values are in bold.
Discussion
This study provided evidence that variants of POMC might be common genetic risk factors for three substance dependence traits (AD, CD, and/or OD). We discuss below several issues concerning study design, result interpretation, and study limitations.
Selection of Non-coding Variants for Common Disease Study
POMC is conservative in its coding region, presumably because several important hormonal peptides are derived from the same POMC precursor. A single mutation in the POMC coding region may have a detrimental effect on the expression and biological functions of several of these peptides. A few diseases or conditions, such as severe early-onset obesity, adrenal insufficiency, and red hair pigmentation have been attributable to these rare loss-of-function mutations (21). Because variants in the POMC coding regions are rare and they may have a harmful impact on POMC expression or cleavage, it is unlikely that these rare coding variants contribute to the risk of common disorders such as drug or alcohol dependence. Instead, variation in non-coding regions may regulate POMC expression and contribute to the risk for these common disorders. Therefore, we chose five non-coding SNP markers for this study.
Positive Association of A Downstream Variant
Among the five POMC SNPs, rs1866146, which is located in the downstream region, showed the most significant result. Although this variant does not change POMC peptide sequence or modify subsequent protein cleavage, given its positive association with substance dependence (this study) and obesity traits (20), we cannot exclude the possibility that it regulates POMC transcription. By querying the sequence harboring rs1866146 against the Transcription Element Search System (TESS) database TRANSFAC v6.0 (http://www.cbil.upenn.edu/tess), we found that the core binding site (GCCTC) of transcription factor T-Ag harbors the minor allele (C) [rather than the major allele (T)] of rs1866146. TRANSFAC analysis is considered provisional since prediction of transcription factor binding sites is far from full-proof.
There is evidence that pituitary POMC expression in mammals depends on the activity of several distinct binding sites and transcription factors (38). The sequence harboring rs1866146 may be contained in one of these transcription factor binding sites. Additionally, even though a positive association signal in the 3′ region of POMC (represented by rs6713532 and rs1866146) was observed, we cannot exclude the possibility that the association may be driven by a functional polymorphism which is in close LD with these two non-coding variants.
Family-based and Case-control Studies in Two Populations
Given the advantages and difficulties of family- and case-control-based studies, we employed both the affected sibling pair (ASP) study approach (one type of family-based studies) and the case-control study approach, which we believe to be complementary, in the present study. Additionally, as allele frequency of variants often differs markedly in populations with different ancestral origins and a disease allele in one population may not exert any effect on the disease in another population, we examined the association of POMC variants with alcohol or drug dependence in both African Americans (AAs) and European Americans (EAs). In other words, four independent sets of samples (both AA and EA family samples and both AA and EA case-control samples) were included in this study.
The results from our case-control samples supported the results from our family samples, but were generally stronger, possibly because the case-control sample had higher power than the family sample to detect SNP effects. Family-based analyses revealed an association of rs6719226 with OD in AA families, and rs6713532 with CD in EA families (Table 3). Similarly, case-control analyses demonstrated a trend for an association between rs6719226 and OD in AAs, and a significant association of rs6713532 with AD or CD in EAs (Table 4). Moreover, rs1866146 was strongly associated with substance dependence traits in both AAs and EAs, even after the conservative Bonferroni correction. Rs6713532 was in a moderate LD with rs1866146 in EA samples. It is unknown whether rs6713532 is a separate disease variant or the positive association generated from this marker is due to its LD with the downstream variant rs1866146. Further studies are warranted to clarify whether the effects of these two variants are related or independent.
Additionally, by haplotype analyses, we only found positive association results in our EA case-control samples (Table S2), but not in our AA and EA family samples (Table S1) or AA case-control samples (Table S2). There may be two possible explanations. One is that the five markers were not in compete LD (see Figure 1), so haplotype analysis (using the five markers) results cannot completely reflect single-marker analysis results (and have reduced power compared to those analyses because of the multiple haplotypes that must be considered in the context of dilution of results from whatever SNP or SNPs are actually driving the result). Another is that, in AA family or AA case-control samples, the association between POMC variants and substance dependence might be only at the individual marker level. Thus, the interactive effect of the five markers on substance dependence in AA population was not detectable by haplotype analyses.
Study Limitations and Solutions
There are three main issues that limit the conclusion that can be drawn from the present study, though an effort was made to overcome all of these limitations. First, there were relatively few informative families in both our AA and EA family samples. This is due to missing genotype data for parents in some families and the low allele frequency of several POMC markers, and this reflects the lower power of ASP-based samples to detect SNP effects when analyzed by FBAT, compared to case-control samples. To overcome this difficulty and to verify the results from family-based studies, we replicated the study with both AA and EA case-control samples. Our case-control studies partially supported the results from our family-based studies, and furthermore, provided new evidence of allelic association between POMC and substance dependence. Second, in our case-control samples, there were more male cases and younger controls. To address this issue, we re-analyzed the data using backward stepwise logistic regression in which confounding factors (age and sex) were considered. The results obtained supported the findings from marker or haplotype association analyses. Third, case-control studies are vulnerable for Type I errors due to population stratification. To address this issue, we applied complementary family- and case-control-based approaches. Additionally, a panel of 38 AIMs were available for all family members (27) and a majority (96.1%) of the unrelated subjects (Figure 2), making it possible to classify these individuals as either genetic ‘AAs’ or genetic ‘EAs’. Thus, the positive results from our case-control studies were unlikely to have been driven by population stratification. However, this set of 38 AIMs may not have sufficient power to detect subtle allele frequency differences occurring within the same population group (e.g., allele frequency differences between Northern and Southern Europeans, or East and West Africans). Of particular note is the consistency of the findings from both family- and population-based approaches, which provide convergent validation of the association findings. Finally, the issue of cumulative multiple testing needs to be considered, because many of our samples (our family samples and SCID-III-R or SCID-IV evaluated case-control samples) have been employed in previous studies. It is unclear how this should be addressed; we believe that it would be too stringent to correct the association results obtained in this study by the number of tested genes to date plus the number of tested markers in each gene; markers with minor or moderate effects would be neglected.
We note that two different structured instruments were used in establishing diagnosis, the SCID (two versions, for DSM-III-R and for DSM-IV) and the SSADDA. Strong cross-system agreement for substance use disorders as defined by DSM-III-R, DSM-IV and ICD-10 (the International Classification of Diseases, version 10) has been reported (39). Moreover, the SSADDA can yield reliable DSM-IV diagnoses for a variety of psychiatric disorders, including alcohol and drug dependence (26,40). Therefore, it is unlikely that multiple measures applied in this study led to significant phenotypic heterogeneities.
In summary, both our family- and case-control studies provided evidence of association between POMC and substance dependence. In fact, our positive results stemmed largely from the downstream variant rs1866146, suggesting a potential gene regulation mechanism in the 3′ flanking region. Thus, functional study of this variant is warranted.
Supplementary Material
Acknowledgments
AnnMarie Lacobelle and Gregory Dalton-Kay provided excellent technical assistance. Colin A. Hodgkinson, Ph.D., helped with genotyping by the Illumina GoldenGate Assay methodology. Konstantin Voronin, M.D., Ph.D., and Carolyn Wenner assisted in data collection at the Medical University of South Carolina. Critical database management services were provided by John Farrell. We thank the families and individuals who volunteered to participate in this study and the expert interviewers who phenotyped the participants.
This work was supported in part by funds from the National Institute on Drug Abuse (R01 DA12849, R01 DA12690, K24 DA15105, K24 DA022288 and K99DA022891), the National Institute on Alcohol Abuse and Alcoholism (R01AA11330, P50 AA12870, K08 AA13732, and K24 AA13736), and the National Center for Research Resources (M01 RR06192; University of Connecticut General Clinical Research Center); and the U.S. Department of Veterans Affairs [the National Center for PTSD Research, the VA Medical Research Program and the VA Connecticut-Massachusetts Mental Illness Research, Education and Clinical Center (MIRECC), and VA Research Enhancement Award Program (REAP), and MERIT Program]. It was also partly supported by the Alcoholic Beverage Medical Research Foundation (ABMRF) grant (to H. Zhang).
Footnotes
Financial Disclosures
Although not directly related to this study, Dr. Kranzler has received research support from Ortho-McNeil Pharmaceuticals and Bristol-Myers Squibb Co. and has been a paid consultant for Forest Pharmaceuticals; Alkermes, Inc.; Ortho-McNeil Pharmaceuticals; elbion NV; sanofi-aventis; Solvay Pharmaceuticals; and H. Lundbeck A/S. Dr. Weiss has received research funding from Eli Lilly and Company. The other authors report no biomedical financial interests or potential conflicts of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Bender T, Nagy G, Barna I, Tefner I, Kadas E, Geher P. The effect of physical therapy on beta-endorphin levels. Eur J Appl Physiol. 2007;100:371–382. doi: 10.1007/s00421-007-0469-9. [DOI] [PubMed] [Google Scholar]
- 2.Appleyard SM, Hayward M, Young JI, Butler AA, Cone RD, Rubinstein M, et al. A role for the endogenous opioid beta-endorphin in energy homeostasis. Endocrinology. 2003;144:1753–1760. doi: 10.1210/en.2002-221096. [DOI] [PubMed] [Google Scholar]
- 3.Millington GW. Proopiomelanocortin (POMC): the cutaneous roles of its melanocortin products and receptors. Clin Exp Dermatol. 2006;31:407–412. doi: 10.1111/j.1365-2230.2006.02128.x. [DOI] [PubMed] [Google Scholar]
- 4.Luger TA, Scholzen TE, Brzoska T, Bohm M. New insights into the functions of alpha-MSH and related peptides in the immune system. Ann N Y Acad Sci. 2003;994:133–140. doi: 10.1111/j.1749-6632.2003.tb03172.x. [DOI] [PubMed] [Google Scholar]
- 5.Flik G, Klaren PH, Van den Burg EH, Metz JR, Huising MO. CRF and stress in fish. Gen Comp Endocrinol. 2006;146:36–44. doi: 10.1016/j.ygcen.2005.11.005. [DOI] [PubMed] [Google Scholar]
- 6.Olive MF, Koenig HN, Nannini MA, Hodge CW. Stimulation of endorphin neurotransmission in the nucleus accumbens by ethanol, cocaine, and amphetamine. J Neurosci. 2001;21:RC184. doi: 10.1523/JNEUROSCI.21-23-j0002.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dai X, Thavundayil J, Santella S, Gianoulakis C. Response of the HPA-axis to alcohol and stress as a function of alcohol dependence and family history of alcoholism. Psychoneuroendocrinology. 2007;32:293–305. doi: 10.1016/j.psyneuen.2007.01.004. [DOI] [PubMed] [Google Scholar]
- 8.Harbuz MS, Lightman SL. Responses of hypothalamic and pituitary mRNA to physical and psychological stress in the rat. J Endocrinol. 1989;122:705–711. doi: 10.1677/joe.0.1220705. [DOI] [PubMed] [Google Scholar]
- 9.Jensen JB, Mork A, Mikkelsen JD. Chronic antidepressant treatments decrease pro-opiomelanocortin mRNA expression in the pituitary gland: effects of acute stress and 5-HT(1A) receptor activation. J Neuroendocrinol. 2001;13:887–893. doi: 10.1046/j.1365-2826.2001.00712.x. [DOI] [PubMed] [Google Scholar]
- 10.Moldow RL, Fischman AJ. Cocaine induced secretion of ACTH, beta-endorphin, and corticosterone. Peptides. 1987;8:819–822. doi: 10.1016/0196-9781(87)90065-9. [DOI] [PubMed] [Google Scholar]
- 11.Yang XM, Gorman AL, Dunn AJ, Goeders NE. Anxiogenic effects of acute and chronic cocaine administration: neurochemical and behavioral studies. Pharmacol Biochem Behav. 1992;41:643–650. doi: 10.1016/0091-3057(92)90386-t. [DOI] [PubMed] [Google Scholar]
- 12.Roth-Deri I, Zangen A, Aleli M, Goelman RG, Pelled G, Nakash R, et al. Effect of experimenter-delivered and self-administered cocaine on extracellular beta-endorphin levels in the nucleus accumbens. J Neurochem. 2003;84:930–938. doi: 10.1046/j.1471-4159.2003.01584.x. [DOI] [PubMed] [Google Scholar]
- 13.Zalewska-Kaszubska J, Gorska D, Dyr W, Czarnecka E. Effect of acute administration of ethanol on beta-endorphin plasma level in ethanol preferring and non-preferring rats chronically treated with naltrexone. Pharmacol Biochem Behav. 2006;85:155–159. doi: 10.1016/j.pbb.2006.07.028. [DOI] [PubMed] [Google Scholar]
- 14.Winkler A, Roske I, Furkert J, Fickel J, Melzig MF. Effects of voluntary ethanol ingestion on the POMC gene expression in the rat pituitary and on the plasma beta-endorphin content. Alcohol Alcohol. 1995;30:231–238. [PubMed] [Google Scholar]
- 15.O’Malley SS, Krishnan-Sarin S, Farren C, Sinha R, Kreek MJ. Naltrexone decreases craving and alcohol self-administration in alcohol-dependent subjects and activates the hypothalamo-pituitary-adrenocortical axis. Psychopharmacology (Berl) 2002;160:19–29. doi: 10.1007/s002130100919. [DOI] [PubMed] [Google Scholar]
- 16.Heinz A, Rommelspacher H, Graf KJ, Kurten I, Otto M, Baumgartner A. Hypothalamic-pituitary-gonadal axis, prolactin, and cortisol in alcoholics during withdrawal and after three weeks of abstinence: comparison with healthy control subjects. Psychiatry Res. 1995;56:81–95. doi: 10.1016/0165-1781(94)02580-c. [DOI] [PubMed] [Google Scholar]
- 17.Kiefer F, Jahn H, Schick M, Wiedemann K. Alcohol self-administration, craving and HPA-axis activity: an intriguing relationship. Psychopharmacology (Berl) 2002;164:239–240. doi: 10.1007/s00213-002-1255-3. [DOI] [PubMed] [Google Scholar]
- 18.Esel E, Sofuoglu S, Aslan SS, Kula M, Yabanoglu I, Turan MT. Plasma levels of beta-endorphin, adrenocorticotropic hormone and cortisol during early and late alcohol withdrawal. Alcohol Alcohol. 2001;36:572–576. doi: 10.1093/alcalc/36.6.572. [DOI] [PubMed] [Google Scholar]
- 19.Van Ree JM. Role of pituitary and related neuropeptides in alcoholism and pharmacodependence. Prog Neuropsychopharmacol Biol Psychiatry. 1986;10:219–228. doi: 10.1016/0278-5846(86)90076-x. [DOI] [PubMed] [Google Scholar]
- 20.Sutton BS, Langefeld CD, Williams AH, Norris JM, Saad MF, Haffner SM, et al. Association of proopiomelanocortin gene polymorphisms with obesity in the IRAS family study. Obes Res. 2005;13:1491–1498. doi: 10.1038/oby.2005.180. [DOI] [PubMed] [Google Scholar]
- 21.Krude H, Biebermann H, Luck W, Horn R, Brabant G, Gruters A. Severe early-onset obesity, adrenal insufficiency and red hair pigmentation caused by POMC mutations in humans. Nat Genet. 1998;9:155–157. doi: 10.1038/509. [DOI] [PubMed] [Google Scholar]
- 22.Tsuang MT, Lyons MJ, Eisen SA, Goldberg J, True W, Lin N, et al. Genetic influences on DSM-III-R drug abuse and dependence: a study of 3,372 twin pairs. Am J Med Genet. 1996;67:473–477. doi: 10.1002/(SICI)1096-8628(19960920)67:5<473::AID-AJMG6>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 23.Xuei X, Flury-Wetherill L, Bierut L, Dick D, Nurnberger J, Jr, Foroud T, et al. The opioid system in alcohol and drug dependence: family-based association study. Am J Med Genet B Neuropsychiatr Genet. 2007;144:877–884. doi: 10.1002/ajmg.b.30531. [DOI] [PubMed] [Google Scholar]
- 24.Racz I, Schurmann B, Karpushova A, Reuter M, Cichon S, Montag C, et al. The opioid peptides enkephalin and beta-endorphin in alcohol dependence. Biol Psychiatry. 2008;64:989–997. doi: 10.1016/j.biopsych.2008.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gelernter J, Yu Y, Weiss R, Brady K, Panhuysen C, Yang BZ, et al. Haplotype spanning TTC12 and ANKK1, flanked by the DRD2 and NCAM1 loci, is strongly associated to nicotine dependence in two distinct American populations. Hum Mol Genet. 2006;15:3498–3507. doi: 10.1093/hmg/ddl426. [DOI] [PubMed] [Google Scholar]
- 26.Pierucci-Lagha A, Gelernter J, Feinn R, Cubells JF, Pearson D, Pollastri A, et al. Diagnostic reliability of the Semi-structured Assessment for Drug Dependence and Alcoholism (SSADDA) Drug Alcohol Depend. 2005;80:303–312. doi: 10.1016/j.drugalcdep.2005.04.005. [DOI] [PubMed] [Google Scholar]
- 27.Gelernter J, Panhuysen C, Weiss R, Brady K, Hesselbrock V, Rounsaville B, et al. Genomewide linkage scan for cocaine dependence and related traits: significant linkages for a cocaine-related trait and cocaine-induced paranoia. Am J Med Genet B Neuropsychiatr Genet. 2005;136:45–52. doi: 10.1002/ajmg.b.30189. [DOI] [PubMed] [Google Scholar]
- 28.Zhang H, Kranzler HR, Yang BZ, Luo X, Gelernter J. The OPRD1 and OPRK1 loci in alcohol or drug dependence: OPRD1 variation modulates substance dependence risk. Mol Psychiatry. 2008;13:531–543. doi: 10.1038/sj.mp.4002035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 30.Shi MM, Myrand SP, Bleavins MR, de la Iglesia FA. High throughput genotyping for the detection of a single nucleotide polymorphism in NAD(P)H quinone oxidoreductase (DT diaphorase) using TaqMan probes. Mol Pathol. 1999;52:295–299. doi: 10.1136/mp.52.5.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Luo X, Kranzler HR, Zuo L, Yang BZ, Lappalainen J, Gelernter J. ADH4 gene variation is associated with alcohol and drug dependence: results from family controlled and population-structured association studies. Pharmacogenet Genomics. 2005;15:755–768. doi: 10.1097/01.fpc.0000180141.77036.dc. [DOI] [PubMed] [Google Scholar]
- 32.O’Connell JR, Weeks DE. PedCheck: a program for identification of genotype incompatibilities in linkage analysis. Am J Hum Genet. 1998;63:259–266. doi: 10.1086/301904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Horvath S, Xu X, Laird NM. The family based association test method: strategies for studying general genotype--phenotype associations. Eur J Hum Genet. 2001;9:301–306. doi: 10.1038/sj.ejhg.5200625. [DOI] [PubMed] [Google Scholar]
- 34.Horvath S, Xu X, Lake SL, Silverman EK, Weiss ST, Laird NM. Family-based tests for associating haplotypes with general phenotype data: application to asthma genetics. Genet Epidemiol. 2004;26:61–69. doi: 10.1002/gepi.10295. [DOI] [PubMed] [Google Scholar]
- 35.Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Falush D, Stephens M, Pritchard JK. Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics. 2003;164:1567–1587. doi: 10.1093/genetics/164.4.1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu K, Muse SV. PowerMarker: an integrated analysis environment for genetic marker analysis. Bioinformatics. 2005;21:2128–129. doi: 10.1093/bioinformatics/bti282. [DOI] [PubMed] [Google Scholar]
- 38.Therrien M, Drouin J. Pituitary pro-opiomelanocortin gene expression requires synergistic interactions of several regulatory elements. Mol Cell Biol. 1991;11:3492–3503. doi: 10.1128/mcb.11.7.3492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rounsaville BJ, Bryant K, Babor T, Kranzler H, Kadden R. Cross system agreement for substance use disorders: DSM-III-R, DSM-IV and ICD-10. Addiction 1993. 1993;88:337–348. doi: 10.1111/j.1360-0443.1993.tb00821.x. [DOI] [PubMed] [Google Scholar]
- 40.Pierucci-Lagha A, Gelernter J, Chan G, Arias A, Cubells JF, Farrer L, et al. Reliability of DSM-IV diagnostic criteria using the semi-structured assessment for drug dependence and alcoholism (SSADDA) Drug Alcohol Depend. 2007;91:85–90. doi: 10.1016/j.drugalcdep.2007.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.