Abstract
Introduction
Total volume of distribution (VT) determined by graphical analysis (GA) of PET data suffers from a noise-dependent bias. Likelihood estimation in GA (LEGA) eliminates this bias at the region of interest level (ROI), but at voxel noise levels, the variance of estimators is high, yielding noisy images. We hypothesized that incorporating LEGA VT estimation in a Bayesian framework would shrink estimators towards prior means, reducing variability and producing meaningful and useful voxel images.
Methods
Empirical Bayesian estimation in GA (EBEGA) determines prior distributions using a two-step k-means clustering of voxel activity. Results obtained on 8 [11C]-DASB studies are compared with estimators computed by ROI-based LEGA.
Results
EBEGA reproduces the results obtained by ROI LEGA while providing low-variability VT images. Correlation coefficients between average EBEGA VT and corresponding ROI LEGA VT range from 0.963 to 0.994.
Conclusions
EBEGA is a fully automatic and general approach that can be applied to voxel-level VT image creation and to any modelling strategy to reduce voxel-level estimation variability without pre-filtering of the PET data.
Keywords: Logan, Likelihood, Bayes, Clustering, Voxel, Serotonin
Introduction
Quantitative analysis of a positron emission tomography (PET) activity requires a mathematical model of the underlying physiological process in order to convert the radioactive tracer concentration into important physiological parameters such as the product of receptor density and the binding affinity. In graphical analysis (GA), the linear relationship found in [1] between the transformed concentration of the radioligand in the plasma and in each target brain region allows one to estimate the total volume of distribution (VT) as the slope of this linear relationship. This approach has been widely adopted across a variety of different radiotracers [2–4]. However, straightforward application of linear regression procedures on the transformed data violates the assumptions underlying these methods. In particular, there is a well-known negative bias in the estimation of the slope, and thus of VT, when ordinary least squares (OLS) methods are used to fit a line to the transformed data, and this becomes more pronounced in regions with high noise levels [5].
Due to the importance of this method and its wide spread use, several strategies have been proposed to correct or reduce this bias, which include incorporating a pre-smoothing step [6–8], minimizing the squared perpendicular distance to the regression line [9], or multi-linear analysis [10]. Likelihood estimation in GA (LEGA) [11] incorporates, in the original non-transformed PET domain, the specific assumptions made on the noise inherent in the measurements (i.e. independent Gaussian noise) and gives optimal estimators of the slope parameter based on likelihood theory, thus yielding approximately unbiased estimators. This yields good estimates when applied at the region of interest (ROI) level [12]. Moreover, LEGA has been validated with test-retest data obtained with the radioligand [11C]N, N-dimethyl-2-(2-amino-4-cyanophenylthio) benzylamine ([11C]-DASB) and proved to be the method of choice among competitive approaches, including one- and two-tissue compartment model [13].
LEGA has recently started to be considered by other groups of investigators as a valid alternative to GA [14]. However, so far one potential barrier for the widespread use of LEGA has been the inability to use it in voxel-based modelling approaches. In fact, at the higher noise levels encountered with voxel-based analysis LEGA estimates show a variance too high for routine application (e.g. SPM analysis) yielding noisy images of estimated VT.
Among the methods proposed in the literature, the multi-linear reference tissue model [15] has been demonstrated to allow the rapid generation of parametric images with a relatively small bias compared to kinetic analysis. This approach has been used extensively since its introduction [16–19]. Nevertheless, as with every reference approach, it does not allow for estimation of the VT and requires the existence of a reference region with little or no specific binding that does not differ between groups of interest, which is not available for all radioligands. Studies have been published in which a difference in the reference region has been reported: in [20] the VT of the reference region (the cerebellum) was found to be higher in the controls group thus obviating the possibility of detecting a difference between depressed patients with bipolar disorder and controls when comparing just the non displaceable binding potential (i.e. BPND = (VT – VTref)/VTref, with VTref the VT in the reference region), which is the only outcome measure available when using reference tissue approaches. In [21] it was suggested that reference region approaches cannot be used to detect differences in 5-HT1A receptors between men and women, since the latter present a higher VT compared with men in the reference region, even when a cerebellar subregion devoid of specific binding (i.e. no 5-HT1A receptors) was located.
Recently, a maximum a posteriori (MAP)-based estimation algorithm has been applied to graphical analysis (MEGA) to reduce the variance of the VT images obtained by LEGA by shrinking the estimators of VT towards a prior mean [22]. The results obtained in [22] on both simulated and clinical data suggest that MEGA represents a valid alternative to LEGA, since it improves the signal-to-noise ratio in clinical VT images with a high correlation with results obtained on the ROI level and a relatively small underestimation. However, the selection of the priors proposed in [22], as well as the parameters of the algorithm setting, seem to be bound to the a priori knowledge of the investigated radioligand (i.e. the [11C]SA4503) and are not data dependent.
To overcome this limitation, we propose to estimate VT using LEGA embedded within a Bayesian framework in order to shrink estimators of VT towards a prior mean by determining reasonable prior distributions and weighting the prior information appropriately, thus reducing variability, as done in [22]. Differently from [22], however, we adopt the so-called empirical Bayesian approach, in which the prior information is determined in a fully automated way from the observed data to eliminate reliance of the estimators on subjectively chosen prior distributions. We term the application of this approach “empirical Bayesian estimation in graphical analysis” (EBEGA).
This fully automatic approach is tested on a serotonin transporter radioligand ([11C]-DASB). We hypothesize that EBEGA reduces the outliers found in LEGA VT images while at the same time maintaining agreement with results obtained by using LEGA on the ROI level, thus lowering the barrier to a more widely use of LEGA as a ROI- and voxel-based graphical modelling approach.
Materials and methods
Subjects
Eight healthy volunteers studied with [11C]-DASB were included in this study. The Institutional Review Board of the New York State Psychiatric Institute approved the protocols. Subjects gave written informed consent after an explanation of the study.
PET protocol
Preparation of the radioligand, emission data acquisition and reconstruction, and determination of arterial input indices were obtained as previously described for [11C]-DASB [13, 23]. All emission data were acquired in 3D mode on an ECAT HR+ (Siemens/CTI, Knoxville, TN, U.S.A.) after a 10 minute transmission scan. Emission data were collected for 120 minutes by using 21 frames of increasing duration: 3 × 20 seconds, 3 × 1 minute, 3 × 2 minutes, 2 × 5 minutes, and 10 × 10 minutes post-injection.
Images were reconstructed to a 128 × 128 matrix, with a pixel size of 2.5 × 2.5 mm2. Reconstruction was performed with attenuation and scatter correction using the transmission data. The reconstruction and estimated image filters were Shepp 0.5, with 2.5 mm in full width at half maximum (FWHM); the Z filter was all-pass 0.4, with 2.0 mm in FWHM, and the zoom factor was 4.0, leading to a final image resolution of 5.1 mm in FWHM at the center of the field of view [24].
The un-metabolized fraction data were fitted as described in [25]. The input function was calculated as the product of total plasma counts and fitted parent fraction and was fitted using a sum of three exponentials [25].
Image analysis
Images were analyzed using Matlab Release 2006b (The Mathworks, MA) with extensions to the following open source packages: Functional Magnetic Resonance Imaging of the Brain’s Linear Image Registration Tool (FLIRT) v5.2 [26], Brain Extraction Tool (BET) v1.2 [27] and University College of London’s Statistical Parametric Mapping (SPM5, Wellcome Department of Imaging Neuroscience, London, UK) normalization and segmentation routines. Motion correction was applied and anatomical ROIs were traced on the basis of brain atlases and published reports, as described in [12, 25]. On average, 50 ROIs were considered in each subject, which include the anterior cingulate, amygdala, cingulate, dorsolateral prefrontal cortex, hippocampus, insula, medial prefrontal cortex, occipital, ventral prefrontal cortex, parietal, parahippocampal gyrus, temporal, dorsal caudate, dorsal putamen, entorhinal cortex, midbrain, posterior parahippocampal gyrus, thalamus, and ventral striatum.
Magnetic resonance imaging acquisition and segmentation
A detailed description of magnetic resonance protocol parameters, de-scalping, and image segmentation between grey matter (GM), white matter, and cerebrospinal fluid voxels has been already published for [11C]-DASB [13, 23]. GM voxels are extracted on the basis of GM probability masks (SPM5) with probability threshold of 1%.
Likelihood estimation in GA (LEGA)
The basis for GA is given by the linear relationship between the transformed concentration of the radioligand in the plasma and in the target brain region [1]. For compartmental models exhibiting reversible kinetics, the method by Logan et al. rearranges the plasma Cp(t) radioligand concentration and the tissue time activity curve TAC(t) into a linear relation after the equilibrium point t*:
| (1) |
The slope parameter β in Equation 1 represents the tissue VT, the parameter of interest, while the intercept γ has no physiological meaning.
The simple linear regression model is written as Y = Iγ + xβ + ε, where Y = [Y1, …, Yn]T, I is a n x 1 vector of ones, x = [x1, …, xn] T, and ε = [ε1, …, εn] T. In this model, the error ε is assumed to be Gaussian, additive, uncorrelated, and to affect only the response variables. However, if the quantities in Equation 1 are replaced with their noisy observed counterparts and OLS is then applied, the error is multiplicative and strongly correlated for both x and the response variables. The result of this violation of assumptions is that the estimator is negatively biased for β [5].
LEGA [11, 12] is an estimation technique that operates in the original non-transformed domain by incorporating the specific assumptions made on the noise inherent in the measurements. It is based on a rearrangement of Equation 1 for i = k, k + 1, …, n:
| (2) |
by denoting the idealized “noise-free” data as , with ti = (si−1 + si)/2, and expressing the integral in terms of the values, where si represents the end-point of the ith PET scan frame. If the errors with which all the actual Ri values are observed are assumed to be independent Gaussians (i.e. , i = 1,2,…n), then the maximum likelihood estimator is obtained by minimizing via non-linear optimization algorithms the quantity over all choices of β and γ, where the optimal weights wi should be inversely proportional to the variances of the Ri values (e.g., ). Several methods to determine the time point t* after which Equation 1 holds, which could be applicable to determine k in LEGA, have been proposed [10]. However, in the analysis described in this work, the value of k has been chosen based upon visual inspection of many Logan plots [12, 13] and set to use only the last 8 samples in the estimation, corresponding to t* ≈ 45 minutes after injection.
Empirical Bayesian estimation in GA (EBEGA)
Bayesian estimation is a widely accepted approach to shrink estimates towards a mean determined by a priori knowledge about the parameters to be estimated. This prior information, expressed by a probability density function (PDF), is updated from the observed data Ri, giving rise to a posterior distribution for the parameters that is proportional to the product of the prior and the likelihood of Ri.
Given this posterior PDF, several Bayesian estimators can be defined for β and γ. In particular, the MAP estimates β̂MAP and γ̂MAP are obtained as the values that maximize the posterior PDF. Assuming a Gaussian distribution for both the a priori PDF of β and γ, β̂MAP and γ̂MAP can be expressed as
| (3) |
where W is the diagonal weight matrix of weights wi, i = k, k + 1, …, n, and Σ, β̄ and γ̄ are the a priori covariance matrix and mean of β and γ, respectively. β̂MAP and γ̂MAP are obtained by minimizing via non-linear optimization algorithms the cost function in Equation 3 over all choices of β and γ (the Gauss-Newton approach was taken, [28, 29]).
In the so-called empirical Bayesian approach, the prior information is determined from the observed data to eliminate reliance of the estimators on subjectively chosen prior distributions. We term the application of this approach “empirical Bayesian estimation in graphical analysis” (EBEGA). The a priori information plays an important role so that it has to be properly chosen for each voxel. In particular, parameters β and γ should be shrunk towards the mean of different priors able to cover the range of kinetic behaviours present across the brain. The quantities Σ, β̄ and γ̄ may be determined by using the information embedded within each voxel raw TAC. This is accomplished in EBEGA in a fully automatic fashion by allocating all voxels to different clusters, determining a prior for each cluster, and then determining β̂MAP and γ̂MAP for each voxel according to the cluster to which it belongs.
The general idea is to create a first set of clusters, which grossly identify the principal different kinetics present across the brain. A second set of smaller sub-clusters is then formed starting from the first ones in order to discriminate between different shapes in the TACs within each original cluster. In the end, the latter are used to form the prior information for each voxel. To achieve this goal, in each study a two-step k-means clustering algorithm [30] is applied to all GM voxels raw TACs. In the first step, a k-means clustering algorithm with K clusters is repeated several times, each time with a new set of initial cluster centroid positions randomly selected from among the TACs; at the end, the solution with the lowest value of the within-cluster sums of point-to-centroid squared Euclidian distances is selected. The number K is automatically determined for each subject by applying a subtractive clustering algorithm to a subset of randomly chosen raw TACs [31]. This algorithm treats each TAC as a potential cluster center, calculates a measure of the likelihood that each TAC would define the cluster center, based on the density of surrounding TACs, and iterates the process until each of the TACs falls within some distance d of a cluster center. Naturally, smaller choices for d tend to give larger values for K, so in order for our results to depend less on a specific value of d, we apply the subtractive clustering algorithms with values for d ranging from 0.2 to 0.5 (see Discussion for details) and take K to be the average of the number of clusters determined each time by the algorithm.
In the second step, the k-means clustering algorithm is applied to each one of the K clusters extracted during the first step to discriminate in detail among the different kinetics composing the cluster. The number of sub-clusters to be extracted is automatically selected for each cluster by using the subtractive clustering algorithm [31] with values for d ranging from 0.2 to 0.5, as in the first step, but with the requirement of a total number of sub-clusters extracted in the subject lower than 100 (see Discussion for details). On average, the number of voxels included in the final subcluster ranges from a minimum of 50 to a maximum of 7000 voxels.
For each sub-cluster, original ROI-based LEGA [11] is then used to estimate β and γ for the (1) average TAC of all the TACs belonging to the voxels within the extracted cluster (with corresponding estimates denoted here β̂mean and γ̂mean); (2) average TAC plus m times the standard deviation (SD) at each time point of all the TACs belonging to the voxels within the extracted cluster (β̂SD+ and γ̂SD+); and (3) average TAC minus m times the SD at each time point of all the TACs belonging to the voxels within the extracted cluster (β̂SD− and γ̂SD−). These estimates represent the basis on which β̄, γ̄ and the diagonal elements of Σ of the a priori Gaussian distribution of β and γ are set for each cluster: β̄ = β̂mean, γ̄= γ̂mean, and , with σβ =min[|β̂mean − β̂SD−|, |β̂mean − β̂SD+|] and σγ= min[|γ̂mean − γ̂SD−|, |γ̂mean − γ̂SD+|], respectively. This can be done for any choice of multiplier m, noting that as m increases, the quantities |β̂mean − β̂SD−|, |β̂mean − β̂SD+|, |γ̂mean − γ̂SD−| and |γ̂mean − γ̂SD+| also increase, thus leading to higher values for σβ and σγ, and ultimately, to less shrinkage. In our application we set m equal to 1.
Once the prior distributions have been determined, the MAP estimation of both β and γ as expressed in Equation 3 is then performed using for each voxel the prior of the sub-cluster to which it belongs.
Outcome measures and performance indices
Different estimates of VT values were computed for comparison:
EBEGA: EBEGA was applied to obtain parametric VT images for all the subjects. Mean, SD, and coefficient of variation (CV) of all estimated VT values within each ROI were computed for each subject.
VOX LEGA: LEGA was applied at the voxel level to obtain parametric VT images for all the subjects. Mean, SD, and coefficient of variation (CV) of all estimated VT values within each ROI were computed for each subject.
ROI LEGA: For each ROI and for each subject, VT values were estimated by modelling the average TAC within the ROI using LEGA.
The results were compared using the linear correlation coefficient r across all ROIs both for all subjects and separately for each subject. To evaluate the possible bias between VOX LEGA and ROI LEGA, and EBEGA and ROI LEGA, respectively, the slope and the intercept of the fitted regression line, with ROI LEGA being the independent variable and the mean of voxel methods the dependent one, were also calculated for each subject.
LEGA at the ROI level was selected as the reference for evaluating the performance of EBEGA at the voxel level since it has been validated in a test-retest reliability paradigm for [11C]-DASB proving to be the method of choice among competitive approaches [13].
Results
A representative case of the parametric images obtained from applying VOX LEGA and EBEGA is displayed in Fig. 1. This representative case was chosen by averaging the r values obtained in each subject in the comparison EBEGA vs. ROI LEGA and selecting the subject with the closest r to the average value. All subsequent analyses are for all regions, all subjects. VOX LEGA (panel A) often results in noisy parametric images unsuitable for subsequent analysis. The corresponding VT images obtained by EBEGA (panel B) show a significant improvement in terms of variance (i.e. decreased variance). In order to quantify these qualitative differences we compare VOX LEGA and EBEGA to our ‘gold standard’, ROI LEGA (Fig 2). The slope for VOX LEGA considering the regression line is 0.999 with a narrow range (0.905 to 1.044) with a mean of 0.971 ± 0.052. This reflects the fact that VOX LEGA estimators are nearly unbiased at a voxel level, although the intercept often differs from zero (0.788 to 4.278) with a mean of 1.930 ± 1.094.
Fig. 1.
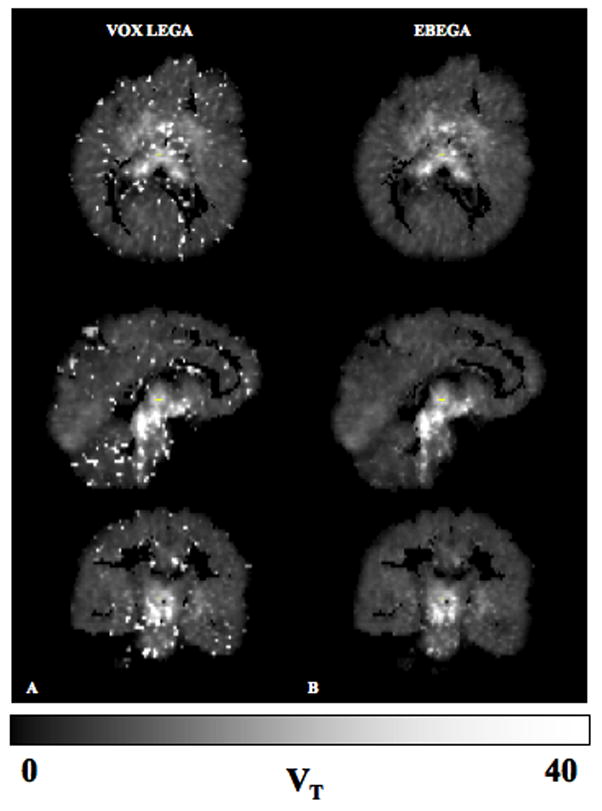
Panels A and B: Estimated VT images obtained by VOX LEGA (A) and EBEGA (B) for a representative case, based on the correlation coefficients between EBEGA and ROI among the 8 considered [11C]-DASB studies.
Fig. 2.
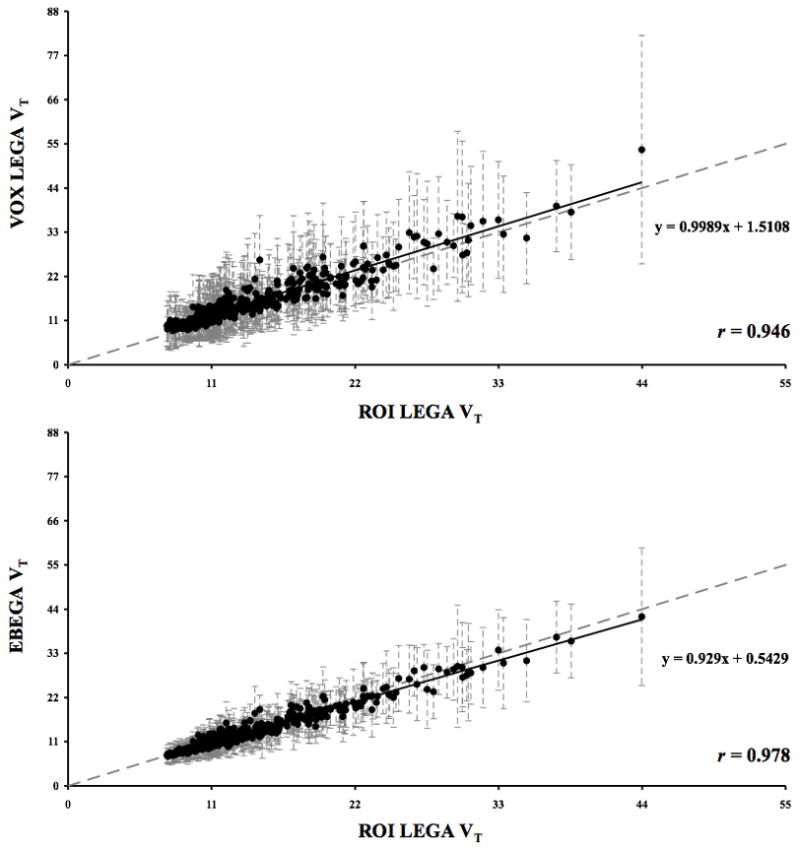
Upper panel: Scatterplot of VOX LEGA (y axis) vs. ROI LEGA (x axis) estimated VT values obtained on all the 392 ROIs considered for the 8 [11C]-DASB studies. Lower panel: Corresponding scatterplot of EBEGA (y axis) vs. ROI LEGA (x axis) estimated VT values. The grey dotted line is the identity line and the black solid line is the fitted regression line. SD bars of VT estimates within each ROI are shown as dashed grey lines in both plots.
EBEGA is associated with a slight reduction in the value of the slope (0.929) obtained considering the regression line. However, EBEGA slope values in each subject are not significantly different from those obtained by VOX LEGA (p = 0.0640; range = 0.906 to 0.976, mean = 0.929 ± 0.026), and intercept values are significantly closer to zero than those obtained by VOX LEGA (p = 0.0039; range = 0.205 to 0.952, mean = 0.555 ± 0.264).
Table 1 reports a summary of the r and CV values in the comparison between VOX LEGA vs. ROI LEGA and EBEGA vs. ROI LEGA. The average CV values of the VT estimated with EBEGA are significantly lower than those obtained with VOX LEGA (p = 0.000013). The improvement in CV by EBEGA is not obtained at the cost of quantitative accuracy; the overall correlation is higher (r = 0.978) than between VOX LEGA and ROI LEGA (r = 0.946). The r values obtained in each subject for EBEGA are significantly higher than those for VOX LEGA, thus suggesting that EBEGA is able to more accurately reproduce the results obtained by ROI LEGA (p = 0.0039). Of note, looking specifically at the midbrain, a region of particular clinical significance, the correlation between EBEGA and ROI LEGA is higher than that between VOX LEGA and ROI LEGA (i.e. correlation across subjects of midbrain ROIs: 0.977 vs. 0.964).
Table 1.
Summary statistics of the r and CV values obtained considering all the ROIs and subjects in the comparison VOX LEGA vs. ROI LEGA and EBEGA vs. ROI LEGA. Min, max, average, and SD refer to the minimum, maximum, average and standard deviation of r and CV values across subjects, respectively. The better performing method according to each criterion is shaded.
| Correlation coefficient (r) |
Average percentage CV |
|||
|---|---|---|---|---|
| VOX LEGA | EBEGA | VOX LEGA | EBEGA | |
| min | 0.890 | 0.963 | 31 | 23 |
| max | 0.970 | 0.994 | 48 | 30 |
| average | 0.941 | 0.976 | 40 | 25 |
| SD | 0.026 | 0.011 | 5.452 | 2.681 |
| global | 0.946 | 0.978 | 40 | 25 |
The decreased variance obtained by EBEGA in the VT images, which is reflected by the CV values reported in Table 1 and the narrower SD bars reported for EBEGA VT estimates in the lower panel of Fig. 2, is in large part due to the elimination of VOX LEGA outliers in the VT distribution. As shown in Fig. 3, where the distributions of VT values estimated by VOX LEGA and EBEGA within three specific ROIs (i.e. amygdala, GM cerebellum, and midbrain) are displayed for the same representative case of Fig. 1, VOX LEGA estimates fall in a wider range of VT values than EBEGA, thus accounting for the outliers that can be detected in VOX LEGA parametric images (Fig. 1, panel A). Considering the VT estimates in the amygdala across all subjects, VOX LEGA shows an average CV value of 41% vs. 28% with EBEGA. The difference is even higher when VT estimates in the GM cerebellum are considered (41% vs. 16%), while results are comparable for midbrain (48% vs. 40%).
Fig. 3.
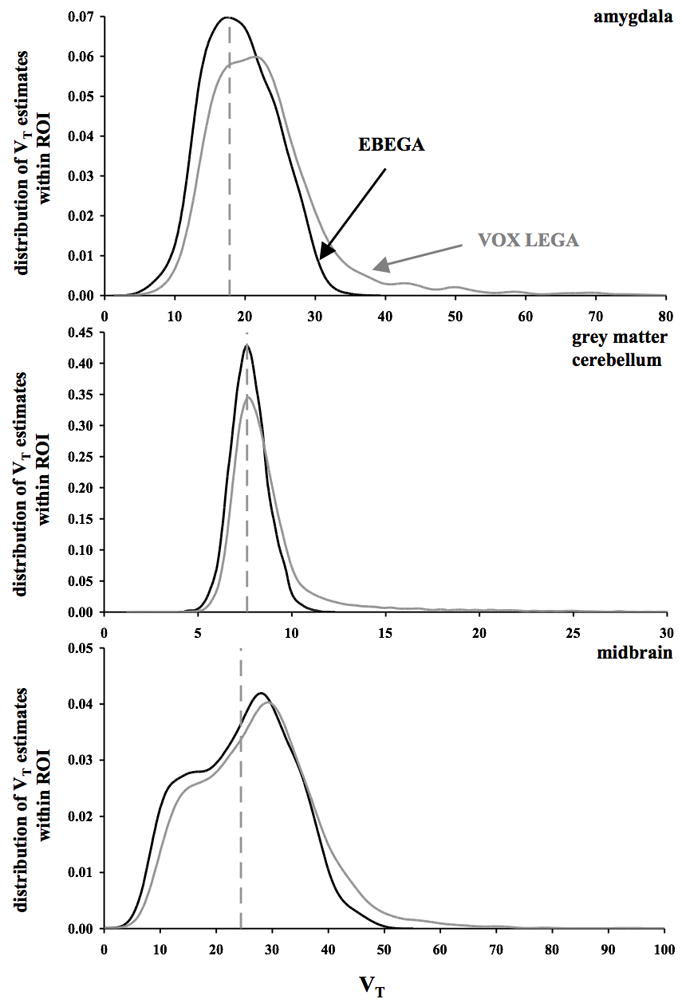
Distribution of VT estimates obtained by VOX LEGA (grey solid line) and EBEGA (black solid line) within three specific ROIs (amygdala, upper, GM cerebellum, middle, and midbrain, lower panel) for the representative case shown in Fig. 1. In each panel, the x axis represents the VT range and the dashed grey vertical line represents the VT estimated by modelling the average TAC for the ROI using LEGA.
The average number of clusters and sub-clusters extracted during the first and second step across all subjects is 12 (range = 7 to 15) and 68 (range = 32 to 100), respectively. To exemplify the effect of the second step of the clustering on the mean of the priors, Fig. 4 shows the maps of the β̂mean value obtained in each voxel after the first and second clusters extraction, respectively, for the representative case of Fig. 1. On average across all subjects, the highest binding cluster β̂mean moves from 29.13 after the first to 60.59 after the second step. A similar trend can be observed with the absolute values of γ̂mean (not shown).
Fig. 4.
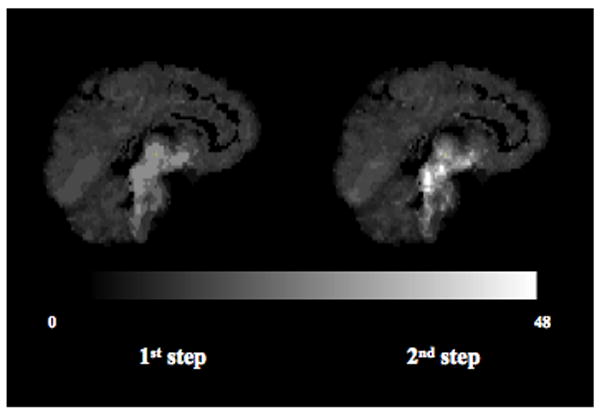
Effect of the second step of clustering on the mean of the Gaussian priors distributions: sagittal view of the β̂mean values that can be obtained in each voxel if the algorithm stops after the first (left) and second (right) clusters extraction, respectively, for the subject shown in Fig. 1.
Discussion
To overcome the noise-dependent bias of the GA and simultaneously produce high quality images, LEGA has been incorporated into a Bayesian framework. The resulting fully automatic EBEGA approach, which implements the MAP estimation and sets the a priori information empirically by applying a two-step k-means clustering to GM voxels raw TACs, has been tested on [11C]DASB data. EBEGA results have been compared to those obtained on a ROI level by LEGA on the basis of correlation coefficients r, and slope and intercept of the fitted regression line.
Some of the assumptions included in the EBEGA algorithm deserve further consideration. First, the number of the clusters to be extracted in both the steps involved in the determination of the a priori information is automatically selected for each subject by applying a subtractive clustering algorithm. This solution is intended to make EBEGA applicable to a wide set of radioligands and/or clinical studies including subjects with diseases, at the same time reducing or eliminating the amount of human supervision required in application. The only parameter that needs to be carefully considered for its impact on the performance of the algorithm is the value of d specifying the cluster center range of influence. In its application inside EBEGA, for each subject the subtractive clustering uses a set of 7 values for d ranging from 0.2 to 0.5. Preliminary investigation outside this range led to values for K which were unreasonably high when d is set below 0.2 (i.e. comparable to the 10% of the total number of GM TACs) and lower than the 5 different tissue types expected, from a physiological point of view, in the GM when d is set over 0.5.
In the application of the subtractive clustering during the second step of the algorithm, a clear trend was observed across all the subjects towards 0.45 as a good choice for d in order to determine the number of sub-clusters for each main cluster. Nevertheless, the EBEGA algorithm was made general by automatically selecting the number of sub-clusters with the only requirement of a total number of sub-clusters extracted in the subject lower than 100. This choice seemed acceptable considering both the kinetics variety present in each subject and the corresponding final performance obtained in VT estimation.
It is however true that experience with a specific radioligand may help in setting a predetermined value for d thus saving computational time, keeping in mind that the principal aim of the clustering is to provide a summary of the varied kinetic behaviours present throughout the brain, to set the a priori information for the MAP estimation, and not to obtain an exhaustive anatomical separation among brain areas. The setting of the priors could also be accomplished by taking more than two steps in the clustering of the voxel TACs. However, given the results obtained for EBEGA in comparison to ROI LEGA, we consider the two-step clustering an acceptable trade-off between computational time and discrimination across different kinetics throughout the brain for all the reported subjects. . In the case of [11C]-DASB in fact, a radioligand that can be described reasonably well with a one-tissue compartment model and shows a narrow range of different kinetics across the brain, to add a further layer of clustering would only result in smaller final subclusters (i.e. with a lower number of voxels) with noisier average TACs and thus potentially leading to noisier VT parametric images. It is also true that, once the Bayesian framework has been defined, alternative prior distributions might be assumed if they prove to be more suitable to the specific data set under analysis. For example, for kinetically richer radioligands, like [11C]-WAY-100635, to add a third step of clustering as well as alternative clustering strategies might be preferable.
Finally, EBEGA sets the a priori mean and diagonal elements of the covariance matrix of the parameter priors for each extracted sub-cluster by applying ROI-based LEGA on the average and the average plus and minus m times the SD of the sub-cluster TACs. Preliminary investigation (not shown) considering the application of LEGA to just the average TAC of each cluster to estimate β̄ and γ̄, and the use of the accuracy of these estimates (i.e. via the Jacobian matrix) to set the diagonal elements of Σ, resulted in priors that were too constraining. Considering the average curve plus and minus m times the SD of the TACs enables one to choose σβ and σγ in a way that reflects the variety of the corresponding TACs family. Increasing the value of m over 1 (not shown) resulted in high values for the diagonal elements of Σ, leading to noisier VT images. A simulated [11C]-DASB mathematical brain phantom created as suggested in [32] was also considered to investigate the effect of the parameter m on the determination of the priors. Specifically, three realizations of the same noisy brain were considered and EBEGA was applied to all of them with three different values of the parameter m: 0.5, 1, and 2. The effect of an increasing value of m in the final outcome measures was a slightly increased SD value in the VT estimates within each ROI as well as a prolonged tail towards higher VT values in the VT estimates distribution, especially for the regions with higher binding. However, considering the percentage difference between the average VT estimated in each ROI by using two different values of m (e.g., in the case of m = 0.5, ), the difference in the estimates obtained by using two values of m is small and ranges between 1.42% and 8.10% for m = 0.5, and 0.37% and 7.20% for m = 2, respectively, across ROIs (i.e. 98 ROIs were considered in the simulated phantoms [32]). These results are consistent with what we would expect from theory: as m increases, the diagonal elements of the covariance matrix of the priors increase thus leading to a more relaxed constraint on the final VT estimates and to potentially noisier parametric VT images. Since the aim of this work was to reproduce the estimates obtained on a ROI level by LEGA, while reducing the variance of VOX LEGA VT images, we consider m = 1 an acceptable trade-off for all the reported studies. One could argue that, in a more complex pathological configuration of cerebral tissue, m = 1 may not be sufficient to describe the variability within a cluster. In such a case, the automated prior determination by itself should be flexible enough to capture the heterogeneity of the different kinetics present in realistic pathological cases.
From a computational point of view, EBEGA does not significantly increase the amount of time needed to obtain VT parametric images with respect to VOX LEGA [12], since the setting of the a priori information before the MAP estimation is reasonably fast.
Conclusion
OLS in graphical analysis can be useful for voxel-based modelling of PET data both because of lower computational time demand and because there are never any convergence problems. However, the violation of assumptions necessary for performing OLS on transformed data leads to biased estimates. On the other hand, to solve this bias by taking into account the nature of the noise in the data and applying more sophisticated estimators can lead to high variability of the estimates. EBEGA has proved to be a reliable fully automatic method able to solve simultaneously the issue of bias and high variability of the estimates while providing low-variability VT images without any pre-filtering of the PET data. Furthermore, the approach adopted inside EBEGA represents a general and flexible method that can be potentially applied to any radioligand and modelling approach to set the prior information from the observed data when Bayesian estimation is used to create parametric images.
Acknowledgments
Funding for studies provided by NARSAD and NIH grants 5 R01 MH040695-17 and 5 P50 MH062185-08
Footnotes
Disclosure/Conflict of Interest
No one of the authors has conflict of interest to declare.
References
- 1.Logan J, Fowler JS, Volkow ND, Wolf AP, Dewey SL, Schlyer DJ, et al. Graphical analysis of reversible radioligand binding from time-activity measurements applied to [N-11C-methyl]-(−)-cocaine PET studies in human subjects. J Cereb Blood Flow Metab. 1990;10(5):740–7. doi: 10.1038/jcbfm.1990.127. [DOI] [PubMed] [Google Scholar]
- 2.Volkow ND, Fowler JS, Wang GJ, Dewey SL, Schlyer D, MacGregor R, et al. Reproducibility of repeated measures of carbon-11-raclopride binding in the human brain. J Nucl Med. 1993;34(4):609–13. [PubMed] [Google Scholar]
- 3.Price JC, Klunk WE, Lopresti BJ, Lu X, Hoge JA, Ziolko SK, et al. Kinetic modeling of amyloid binding in humans using PET imaging and Pittsburgh Compound-B. J Cereb Blood Flow Metab. 2005;25(11):1528–47. doi: 10.1038/sj.jcbfm.9600146. [DOI] [PubMed] [Google Scholar]
- 4.Schuitemaker A, van Berckel BN, Kropholler MA, Kloet RW, Jonker C, Scheltens P, et al. Evaluation of methods for generating parametric (R-[11C]PK11195 binding images. J Cereb Blood Flow Metab. 2007;27(9):1603–15. doi: 10.1038/sj.jcbfm.9600459. [DOI] [PubMed] [Google Scholar]
- 5.Slifstein M, Laruelle M. Effects of statistical noise on graphic analysis of PET neuroreceptor studies. J Nucl Med. 2000;41(12):2083–8. [PubMed] [Google Scholar]
- 6.Feng D, Wang Z, Huang S. A study on statistically reliable and computationally efficient algorithms for generating local cerebral blood flow parametric images with positron emission tomography. IEEE Trans Med Imaging. 1993;12:182–8. doi: 10.1109/42.232247. [DOI] [PubMed] [Google Scholar]
- 7.Feng D, Huang SC, Wang Z, Ho D. An unbiased parametric imaging algorithm for non-uniformly sampled biomedical system parameter estimation. IEEE Trans Med Imaging. 1996;15:512–8. doi: 10.1109/42.511754. [DOI] [PubMed] [Google Scholar]
- 8.Logan J, Fowler JS, Volkow ND, Ding YS, Wang GJ, Alexoff DL. A strategy for removing the bias in the graphical analysis method. J Cereb Blood Flow Metab. 2001;21(3):307–20. doi: 10.1097/00004647-200103000-00014. [DOI] [PubMed] [Google Scholar]
- 9.Varga J, Szabo Z. Modified regression model for the Logan plot. J Cereb Blood Flow Metab. 2002;22:240–4. doi: 10.1097/00004647-200202000-00012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ichise M, Toyama H, Innis RB, Carson RE. Strategies to improve neuroreceptor parameter estimation by linear regression analysis. J Cereb Blood Flow Metab. 2002;22(10):1271–81. doi: 10.1097/01.WCB.0000038000.34930.4E. [DOI] [PubMed] [Google Scholar]
- 11.Ogden RT. Estimation of kinetic parameters in graphical analysis of PET imaging data. Stat Med. 2003;22:3557–68. doi: 10.1002/sim.1562. [DOI] [PubMed] [Google Scholar]
- 12.Parsey RV, Ogden RT, Mann JJ. Determination of volume of distribution using likelihood estimation in graphical analysis: elimination of estimation bias. J Cereb Blood Flow Metab. 2003;23(12):1471–8. doi: 10.1097/01.WCB.0000099460.85708.E1. [DOI] [PubMed] [Google Scholar]
- 13.Ogden RT, Ojha A, Erlandsson K, Oquendo MA, Mann JJ, Parsey RV. In vivo quantification of serotonin transporters using [11C]DASB and positron emission tomography in humans: modeling considerations. J Cereb Blood Flow Metab. 2007;27(1):205–17. doi: 10.1038/sj.jcbfm.9600329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Joshi A, Fessler JA, Koeppe RA. Improving PET receptor binding estimates from Logan plots using principal component analysis. J Cereb Blood Flow Metab. 2008;28:852–65. doi: 10.1038/sj.jcbfm.9600584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ichise M, Liow JS, Lu JQ, Takano A, Model K, Toyama H, et al. Linearized reference tissue parametric imaging methods: application to [11C]DASB positron emission tomography studies of the serotonin transporter in human brain. J Cereb Blood Flow Metab. 2003;23:1096–112. doi: 10.1097/01.WCB.0000085441.37552.CA. [DOI] [PubMed] [Google Scholar]
- 16.Ichise M, Vines DC, Gura T, Anderson GM, Suomi SJ, Higley JD, et al. Effects of early life stress on [11C]DASB positron emission tomography imaging of serotonin transporters in adolescent peer- and mother-reared rhesus monkeys. J Neurosci. 2006;26(17):4638–43. doi: 10.1523/JNEUROSCI.5199-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ichise M, Cohen RM, Carson RE. Noninvasive estimation of normalized distribution volume: application to the muscarinic-2 ligand [(18)F]FP-TZTP. J Cereb Blood Flow Metab. 2008;28(2):420–30. doi: 10.1038/sj.jcbfm.9600530. [DOI] [PubMed] [Google Scholar]
- 18.Yaqub M, Tolboom N, Boellaard R, van Berckel BN, van Tilburg EW, Luurtsema G, et al. Simplified parametric methods for [11C]PIB studies. Neuroimage. 2008;42(1):76–86. doi: 10.1016/j.neuroimage.2008.04.251. [DOI] [PubMed] [Google Scholar]
- 19.Meyer PT, Sattler B, Winz OH, Fundke R, Oehlwein C, Kendziorra K, et al. Kinetic analyses of [123I]IBZM SPECT for quantification of striatal dopamine D2 receptor binding: a critical evaluation of the single-scan approach. Neuroimage. 2008;42(2):548–58. doi: 10.1016/j.neuroimage.2008.05.023. [DOI] [PubMed] [Google Scholar]
- 20.Oquendo MA, Hastings RS, Huang YY, Simpson N, Ogden RT, Hu XZ, et al. Brain serotonin transporter binding in depressed patients with bipolar disorder using positron emission tomography. Arch Gen Psychiatry. 2007;64(2):201–8. doi: 10.1001/archpsyc.64.2.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Parsey RV, Arango V, Olvet DM, Oquendo MA, Van Heertum RL, Mann JJ. Regional heterogeneity of 5-HT1A receptors in human cerebellum as assessed by positron emission tomography. J Cereb Blood Flow Metab. 2005;25(7):785–93. doi: 10.1038/sj.jcbfm.9600072. [DOI] [PubMed] [Google Scholar]
- 22.Shidahara M, Seki C, Naganawa M, Sakata M, Ishikawa M, Ito H, et al. Improvement of likelihood estimation in Logan graphical analysis using maximum a posteriori for neuroreceptor PET imaging. Ann Nucl Med. 2009;23(2):163–71. doi: 10.1007/s12149-008-0226-0. [DOI] [PubMed] [Google Scholar]
- 23.Bélanger MJ, Simpson NR, Wang T, Van Heertum RL, Mann JJ, Parsey RV. Biodistribution and radiation dosimetry of [11C]DASB in baboons. Nucl Med Biol. 2004;31(8):1097–102. doi: 10.1016/j.nucmedbio.2004.09.002. [DOI] [PubMed] [Google Scholar]
- 24.Mawlawi O, Martinez D, Slifstein M, Broft A, Chatterjee R, Hwang DR, et al. Imaging human mesolimbic dopamine transmission with positron emission tomography: I. Accuracy and precision of D(2) receptor parameter measurements in ventral striatum. J Cereb Blood Flow Metab. 2001;21:1034–57. doi: 10.1097/00004647-200109000-00002. [DOI] [PubMed] [Google Scholar]
- 25.Parsey RV, Ojha A, Ogden RT, Erlandsson K, Kumar D, Landgrebe M, et al. Metabolite considerations in the in vivo quantification of serotonin transporters using 11C-DASB and PET in humans. J Nucl Med. 2006;47(11):1796–802. [PubMed] [Google Scholar]
- 26.Jenkinson M, Smith S. A global optimisation method for robust affine registration of brain images. Med Image Anal. 2001;5:143–56. doi: 10.1016/s1361-8415(01)00036-6. [DOI] [PubMed] [Google Scholar]
- 27.Smith S. Fast robust automated brain extraction. Human Brain Mapping. 2002;17:143–55. doi: 10.1002/hbm.10062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Coleman TF, Li Y. On the Convergence of Reflective Newton Methods for Large-Scale Nonlinear Minimization Subject to Bounds. Mathematical Programming. 1994;67(2):189–224. [Google Scholar]
- 29.Coleman TF, Li Y. An Interior, Trust Region Approach for Nonlinear Minimization Subject to Bounds. SIAM Journal on Optimization. 1996;6:418–445. [Google Scholar]
- 30.Spath H. Cluster Dissection and Analysis. In: Goldschmidt J, translator. Theory, FORTRAN Programs, Examples. Ellis Horwood Limited; Chichester, and Wiley, New York: 1985. p. 226. [Google Scholar]
- 31.Chiu S. Fuzzy Model Identification Based on Cluster Estimation. Journal of Intelligent & Fuzzy Systems: Applications in Engineering and Technology. 1994;2(3):269–78. [Google Scholar]
- 32.Rahmim A, Dinelle K, Cheng J, Shilov MA, Segars WP, Lidstone SC, Blinder S, Rousset OG, Vajihollahi H, Tsui BMW, Wong DF, Sossi V. Accurate Event-Driven Motion Compensation in High-Resolution PET Incorporating Scattered and Random Events. IEEE Transactions on Medical Imaging. 2008;27(8):1018–1033. doi: 10.1109/TMI.2008.917248. [DOI] [PMC free article] [PubMed] [Google Scholar]


