Summary
The N-terminal domain of thrombospondin-1 (TSPN-1) mediates the protein’s interaction with (1) glycosaminoglycans, calreticulin, and integrins during cellular adhesion, (2) low-density lipoprotein receptor-related protein during uptake and clearance, and (3) fibrinogen during platelet aggregation. The crystal structure of TSPN-1 to 1.8 Å resolution is a β sandwich with 13 antiparallel β strands and 1 irregular strand-like segment. Unique structural features of the N- and C-terminal regions, and the disulfide bond location, distinguish TSPN-1 from the laminin G domain and other concanavalin A-like lectins/glucanases superfamily members. The crystal structure of the complex of TSPN-1 with heparin indicates that residues R29, R42, and R77 in an extensive positively charged patch at the bottom of the domain specifically associate with the sulfate groups of heparin. The TSPN-1 structure and identified adjacent linker region provide a structural framework for the analysis of the TSPN domain of various molecules, including TSPs, NELLs, many collagens, TSPEAR, and kielin.
Introduction
The thrombospondins (TSPs) comprise a family of extracellular glycoproteins that regulates cellular behavior during tissue genesis and repair (Adams and Lawler, 2004; Bornstein et al., 2004; Bornstein and Sage, 2002). TSP-1 is the most extensively characterized member of the family because it was the first to be identified and is readily purified from human blood platelets. TSP-1 null mice exhibit defects in activation of transforming growth factor β (TGFβ) and wound healing (Bornstein et al., 2004; Lawler and Detmar, 2004; Lawler et al., 1998). In addition, TSP-1 and -2 double null mice have fewer synapses (Christopherson et al., 2005). These diverse functions of TSP-1 are mediated by its interaction with a wide variety of proteins and proteoglycans in the extracellular environment and at the cell surface (Chen et al., 2000). The binding sites for proteins and proteoglycans are distributed throughout the various domains that comprise TSP-1 (Chen et al., 2000). In some cases, similar structural and functional domains are present in the other members of the TSP gene family. All TSP family members, except cartilage oligomeric matrix protein (COMP or TSP-5) have a domain of approximately 200 amino acids at their N terminus. This domain of TSP-1, designated TSPN-1, has been found to (1) bind glucosaminoglycans with high affinity, (2) mediate the uptake and clearance of TSP-1, and (3) have antiadhesive activity. The TSPN domain is also found in neuronal NELL proteins (Kuroda et al., 1999), many collagens (Moradi-Ameli et al., 1994), TSPEAR (Scheel et al., 2002), and kielin (Matsui et al., 2000). In general, this domain is at the N terminus of these large matrix proteins and is followed by an α-helical region that mediates multimerization. In many of these molecules, the TSPN domain appears to be involved in proteoglycan binding.
TSPN-1 is readily cleaved from the intact molecule by proteases. Some proteolytic release of the N-terminal domain occurs within the α granules of platelets before secretion of the protein (Damas et al., 2001). Since several platelet proteins bind to heparin-Sepharose, early purification schemes for TSP-1 sought to separate these proteins from TSP-1 on immobilized heparin. These studies revealed that TSPN-1 binds to heparin with high affinity. The affinity of intact TSP-1 for heparin (Kd = 41 nM) is higher than that of TSPN-1 (Kd =~850 nM) produced by tryptic digestion (Wang et al., 2004). Intact TSP-1 also binds to heparan sulfate, dermatan sulfate, and chondroitin sulfate (Herndon et al., 1999; Merle et al., 1997). The dermatan sulfate binding site has been mapped to amino acids 61–95 of TSPN-1 and is involved in the interaction of TSP-1 with decorin (Merle et al., 1997). In addition to decorin, TSP-1 reportedly binds to syndecan-1, -3, and -4, perlecan, and cerebroglycan (Ferrari do Outeiro-Bernstein et al., 2002; Herndon et al., 1999).
The N-terminal domains of TSP-1 and -2 mediate their uptake and clearance (Wang et al., 2004). This process appears to involve a proteoglycan and LRP. LRP is a member of the low-density lipoprotein (LDL) receptor family, and very low-density lipoprotein (VLDL) receptor can mediate TSP-1 uptake in LRP-deficient cells (Mikhailenko et al., 1997). The LRP binding site is included in the first 90 amino acids of TSPN-1 (Wang et al., 2004). Since matrix metalloproteases (MMP) bind to TSP-1 and -2, they can be taken up along with TSP-1. This process has been shown to be important for the regulation of extracellular MMP activity (Yang et al., 2001).
Various cell types attach and form focal adhesion on fibronectin. These structures are disrupted when TSP-1 or a synthetic peptide that contains amino acids 17–35 of TSPN-1 are added to the cells (Murphy-Ullrich et al., 1993). The binding of TSP-1 to cell surface calreticulin results in the recruitment of LRP and heterotrimeric G protein to the complex (Orr et al., 2002). This complex stimulates phosphoinositide 3-kinase, focal adhesion kinase, and ERK signaling and leads to a disruption of focal adhesions. The interaction of TSP-1 with calreticulin has recently been found to mediate T cell motility (Li et al., 2005).
To better understand the function of TSPN-1, we have determined the structures of TSPN-1 and its complex with a synthetic pentameric heparin (Arixtra) by X-ray crystallography. In this paper, we report the globular β sandwich structure of TSPN-1 that can be classified as a member of the concanavalin A-like lectins/glucanases superfamily and has some unique features. We also provide data on how TSPN-1 binds to heparins, and we review binding of some other proposed ligands of TSPN-1. We also discuss the relevance of the TSPN-1 structure to the TSPN domains of other TSP family members and related TSPN-containing proteins.
Results
The Structure of TSPN-1
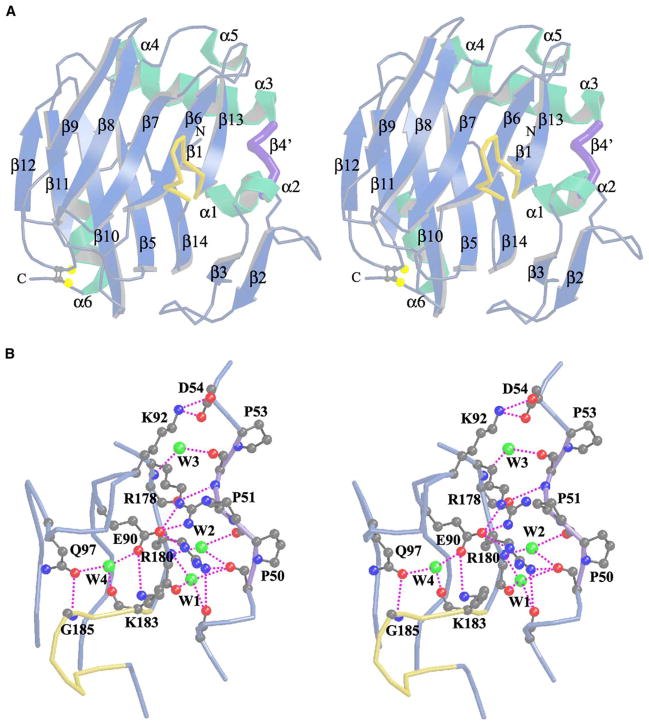
The globular TSPN-1 domain has a β sandwich structure (Figure 1A) and can be classified as a member of the concanavalin A-like lectins/glucanases superfamily according to the Structural Classification of Protein (SCOP) (Murzin et al., 1995). A structure in this superfamily generally consists of two antiparallel β sheets, a concave front sheet, and a convex back sheet with 12–14 strands in total. The concave sheet is the hallmark of the molecules in this superfamily, of which many members bind carbohydrates through a cleft-like motif on the front sheet. The TSPN-1 domain has a somewhat different architecture than the typical β sandwich structure, consisting of 13 antiparallel β strands and 1 irregular strand-like segment (colored in purple) (Figure 1A). This irregular structural segment (including residues P50PVP) forms the right edge of the domain and is a unique feature of the N-terminal region of TSPN-1 (Figure 1B). This proline-rich motif occupies a position of a β strand that is conserved in all other similar β sandwich structures. The replaced β strand is generally anti-parallel to strand β13 and forms part of a jellyroll-like structure together with strands β3, β5, β6, β7, β10, β13, and β14. In many similar β sandwich structures, a jellyroll-like conformation is formed by eight β strands on the right side of the domain. In TSPN-1, the irregular structural segment (designated as β4′ hereafter) shifts away from strand β13 and makes only one direct main chain-to-main chain hydrogen bond with the β13 strand. In addition, several water-mediated hydrogen bonds between strands β13 and β4′ are observed (Figure 1B). Three of them are conserved and well defined in the native structure and the TSPN-1/Arixtra complex discussed below. The irregularity of the β4′ strand makes the already imperfect jellyroll-like structure less defined in TSPN-1. On the left side of the domain, strands of both β sheets are in a more regular up-and-down topology.
Figure 1. The Structure of TSPN-1.
(A) A stereoview of a ribbon diagram of the structure of TSPN-1. The β strands and α helices are colored in cyan and green, respectively. The irregular proline-rich strand-like segment discussed in the text is labeled as β4′ and is highlighted in purple. The sole disulfide bond (yellow) is drawn in ball-and-stick representation. The jar handle-like motif after the β13 strand is highlighted in yellow. The N and C termini are labeled as N and C, respectively. The molecule is oriented so that its concave β sheet or front sheet faces the reader. This figure was prepared with the program MOLSCRIPT (Kraulis, 1991).
(B) A stereoview of the Cα tracing diagram of the front-sheet, right-side strands of TSPN-1. The diagram displays most of the interactions involved with the irregular strand-like segment β4′ colored in purple, interactions with strand β13 and the following jar handle-like motif colored in yellow, as well as a few interactions from strands β6 and β7 and the loop between them. Three water molecules between β4′ and β13 are colored green and are labeled as W1, W2, and W3. They are conserved in both native and TSPN-1/Arixtra structures and are well defined in electron densities. Another water molecule, W4, potentially involved in stabilizing the jar handle motif conformation is also drawn in green.
There are six α helices in TSPN-1 (Figure 1A). They are located either between β strands or at the C-terminal region. Among these helices, α3, crossing over the top of the two β sheets, is the most prominent. Parallel to α3 are two connected short helices, α4 and α5. Two residues between them, R171 and D172, noticeably protrude upward (not shown in the figure). The interactions of all three helices (α3, α4, and α5) with the β sheets are predominantly hydrophobic. We cannot identify a calcium binding site on the top of the molecule like that observed in laminin G-like domain structures (Rudenko et al., 2001). The α6 helix is located at the C terminus and runs parallel to the β strands (Figure 1A). A disulfide bond is formed between C214 at the end of the α6 helix and C153 in the β11-β12 loop. This disulfide bond brings the C terminus into close proximity to the rest of the TSPN-1 domain.
A Dali structure homolog search (http://www.ebi.ac.uk/dali) gave many hits from the concanavalin A-like lectins/glucanases superfamily. The top hits (Z > 10) include the leech intramolecular trans-sialidase (PDB: 2SLI), serum amyloid p component (SAP) (PDB: 1SAC), tetanus neurotoxin (PDB: 1A8D), and calnexin (PDB: 1JHN). However, in these structure alignments, only about 72.6%–81.7% of the residues from TSPN-1 can be aligned. Most of them are from the middle strands of the two β sheets. The strands along the edges, especially on the right edges, vary from structure to structure, showing the complex topologies of the members in this superfamily. Carbohydrates bind to a cleft-like motif in the concave front sheet of the lectins. In TSPN-1, the cleft-like motif is partially covered on the right side by the loop between the β13 and β14 strands (Figure 1A). The cleft-like motif is further obscured by side chains from the β6 (E90) and β7 strands (Q97), which form hydrogen bonds with the loop between the β13 and β14 strands to stabilize it (Figure 1B).
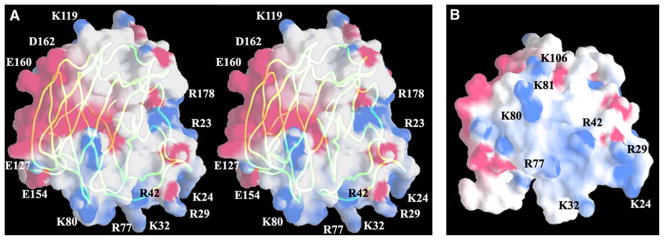
Calculation of the electrostatic potential of TSPN-1 identifies a major positively charged surface patch at the bottom of the domain (Figure 2). A cluster of basic residues including R29, K32, R42, R77, K80, K81, and K106 contributes to this extended positively charged region (Figure 2B). A smaller patch of positive charge on the top of TSPN-1 includes R65, K68, and R171. Most of these basic residues have their side chains projecting into the solvent. Whereas these residues are well separated in the primary sequence, they congregate to form potential heparin binding sites as discussed below. On the right side of the domain, residues K92, R178, R180, and K183 form another cluster of basic amino acids. However, most of these residues are involved in intramolecular salt bridges and/or hydrogen bonds and may not be available for heparin binding (Figure 1B).
Figure 2. Electrostatic Potential Surface Representation of TSPN-1.
(A) A stereoview of TSPN-1 with the major positively (blue) and negatively (red) charged residues labeled. A backbone worm is embedded to show the orientation of the molecule and the positions of the major charged residues. Besides the positively charged patch at the bottom of the domain, there is a negatively charged patch around the left edge and the back of TSPN-1, where fibrinogen reportedly binds.
(B) A view of the positively charged surface patch and the positions of the residues, which may contribute to heparin binding on the bottom of TSPN-1. This site is referred to as the major heparin binding site in the text. The orientation of the molecule shown here is related to the one in (A) by a −60° rotation around the horizontal axis.
These figures were prepared with the program GRASP (Nicholls et al., 1991).
The Structure of TSPN-1/Arixtra
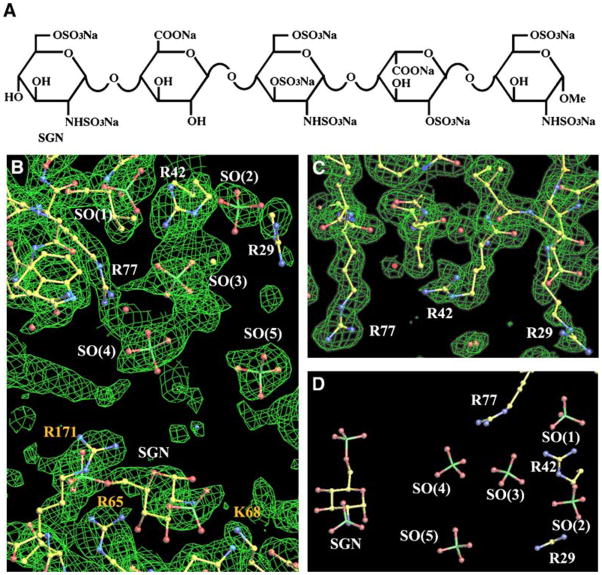
Arixtra (fondaparinux sodium) is a synthetic, modified pentameric heparin (Bauer, 2003) (Figure 3A). It binds to antithrombin III (ATIII) and potentiates the neutralization of Factor Xa. In this study, we used Arixtra as a well-characterized, homogeneous heparin species for co-crystalization with TSPN-1 to study heparin binding.
Figure 3. Arixtra Binding in the TSPN-1/Arixtra Complex.
(A) Chemical structure of Arixtra (fondaparinux sodium), C31H43N3Na10O49S8.
(B) A simulated annealing omit map (2Fo − Fc at the 1.0σ contour level and colored in green) around the major heparin binding site and the partially disordered Arixtra molecule. All non-protein atoms have been removed from the simulated annealing refinement and map calculation. A SGN unit, which binds to the top of a symmetry-related molecule (see text), is labeled in yellow. The density in the middle on the left side is from residue R23 of another symmetry-related molecule. This residue is on the flexible loop (G20AARKG), which changes conformation in the TSPN-1/Arixtra complex. It seems to be involved in an interaction with Arixtra in the molecular packing of the complex, suggesting that the crystal packing rather than heparin binding may induce the conformational changes in the loop.
(C) The major heparin binding site in unligated native TSPN-1 with the difference electron density map (2Fo − Fc) contoured at 1.0σ and colored in green.
(D) The partially disordered Arixtra molecule and the three arginines from the major heparin binding site. In this drawing, the TSPN-1/Arixtra model was rotated about 90° for comparison of the partial Arixtra model with the chemical structure of Arixtra in (A).
In the TSPN-1/Arixtra complex, the structure of TSPN-1 is very similar to that of its native unligated form. A superposition of the two structures gives a root-mean-square deviation (rmsd) of 0.69 Å. The major deviation between the two structures is found in the position of the loop (composed of residues G20AARKG) between the α1 helix and the β2 strand, which is not well-defined by electron densities in the TSPN-1 native structure. If these six residues are not included in the superposition, the resulting rmsd is 0.45 Å. The superposition suggests that there are no significant conformational changes in TSPN-1 due to the association with Arixtra except for this loop, which is close to the major heparin binding site and has been suggested to be one of the GAG binding sites of TSPN-1 (Lawler et al., 1992). The pentameric and polycanionic nature of Arixtra renders the molecule rather flexible with multiple potential binding sites. In the TSPN-1/Arixtra complex structure, either owing to the partial disorder or multiple binding modes, Arixtra does not appear as a well-defined molecule. Nevertheless, bulky and sometimes clustered electron densities that surround residues R29, R42, and R77 in the large positively charged region at the bottom of TSPN-1 clearly represent the bound sulfate groups that are attached to Arixtra’s different carbohydrate rings (Figures 2 and 3B). We have assigned this area as the major heparin binding site. These densities can’t be interpreted as water molecules or as sulfate groups from the HEPES buffer, because both the native TSPN-1 and TSPN-1/Arixtra complex crystals were grown in identical buffer conditions. For comparison, Figure 3C shows the same region of the native TSPN-1 structure, where there are no significant electron densities observed at the assigned major heparin binding site. In the five sulfate groups that have been modeled into the complex structure, three of them, SO(1), SO(2), and SO(3), surround residue R42. This arginine has well-defined electron densities in both native and complex structures. Its cationic guanidinium group changes its orientation in the complex structure such that it is positioned in the center of the triangle formed by the three surrounding anionic sulfate groups. The side chain of residue R29 undergoes a large conformational change, and it is possibly being pulled by SO(2). Residue R77 also appears to have a small conformational change to make a direct interaction with SO(4). SO(5) seems to be stabilized by the interaction with a symmetry-related molecule (not shown in the figure).
The densities for two more SO groups, resembling the fifth unit of Arixtra or a SGN (N, O6-disulfo-glucosamine) unit, are found to be associated with residues R65, K78, and R171 at the top of the molecule. These residues were thought to form a second minor heparin binding site, and their association with the putative SGN unit is shown in the bottom part of Figure 3B. However, from the molecular packing in the crystal, the SGN unit and the five SO groups described earlier seem to be from one single Arixtra molecule (Figure 3D) based on the intermolecular spacing and the dimension of Arixtra (Figure 3A). Thus, one Arixtra molecule binds to one TSPN-1 at the major binding site and also interacts with a symmetry-related TSPN-1 at the minor binding site. Since dimerization of TSPN-1 by Arixtra has not been observed in solution, the basic triplet R65/K78/R171 may form a weak opportunistic binding site that helps to pack TSPN-1 molecules into a crystal lattice during crystallization.
Discussion
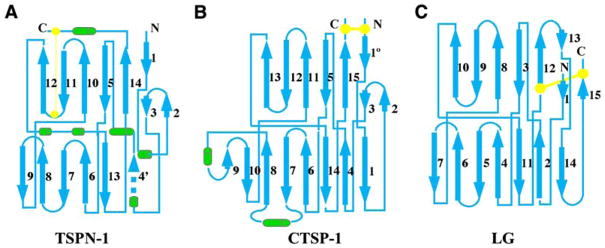
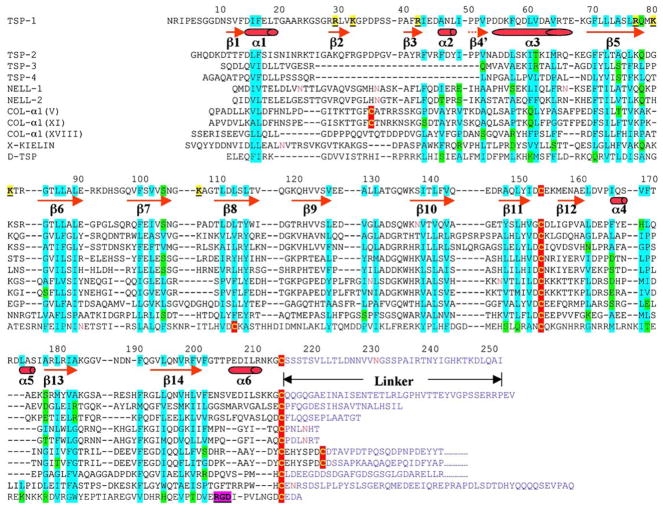
TSPN has been classified as one of the unique modules in extracellular proteins (Bork et al., 1996). It was later predicted to have a jellyroll-like fold as laminin G-like domain (LG), similar to that of pentraxin based on the hydrophobic residue distribution patterns in their conserved regions (Beckmann et al., 1998). Although generally predicted as an antiparallel β sandwich structure for the conserved region, the correct architecture of the globular TSPN-1 has only now been revealed in this study. TSPN-1 has a distinct topology, especially in its disulfide bond location and its N- and C-terminal regions in which TSPN-1 differs from LG, pentraxin, and the other members of the LNS (Laminin A G-domain/Neurexin/Sex hormone binding globulin [SHBG]) repeat family (Rudenko et al., 2001) (Figure 4). Within the concanavalin A-like lectins/glucanases superfamily of the SCOP system, the strand organization of TSPN-1 (Figure 4A) instead is more like that of hydrolases (i.e., trypanosoma rangeli sialidase [PDB: 1MZ5]), lectins (i.e., griffonia simplicifonia [PDB: 1LED]), neurotoxin (i.e., tetanus toxin [PDB: 1A8D]), and calnexin (PDB: 1JHN). In these structures, the N-terminal portion contributes the right-edge strands (Figures 1 and 4A). Thus, the TSPN domain defines a β sandwich structure that is distinct from the laminin G domain. The structure presented here and sequence alignment algorithms enable us to examine other TSPN-containing proteins. Figure 5 shows a TSPN sequence alignment of human TSP-1, TSP-2, TSP-3, TSP-4, Drosophila TSP, NELL-1, and NELL-2 (Kuroda et al., 1999), some collagens (Moradi-Ameli et al., 1994), and kielin (Matsui et al., 2000) based on the structure of TSPN-1. Whereas the sequence identity of the TSPN is relatively low for these proteins, the hydrophobicity pattern of key positions that define the β strands of TSPN-1 is similar in all of the proteins. In addition, the positions of the two cysteine residues (e.g., C153 and C214 in TSPN-1) that form a disulfide bond are conserved. There are additional disulfide bonds in the TSPN domain of some collagens. These data imply that all of these proteins probably have similar β sandwich structures in their TSPN domains. Within the TSP family, the major difference between TSP-1 and -2, and TSP-3 and -4 is the presence of sequence gaps in the latter two proteins within their TSPNs. The missing sequences in TSP-3 and -4 suggest that they are likely to lack the edge β strands 2 and 3, assembling a fold very much like the intramolecular trans-sialidase (PDB: 2SLI). TSPEAR (Scheel et al., 2002) is not included in the sequence alignment. This protein may represent the first TSPN-containing protein that exists as a monomer.
Figure 4. The Topology of TSPN-1.
The topology of (A) TSPN-1 is compared with that of (B) CTSP-1 (the C-terminal domain of TSP-1) (Kvansakul et al., 2004) and (C) a typical LG (Laminin A G-domain [LG]/Neurexin/Sex hormone binding globulin [SHBG]) repeat (Rudenko et al., 2001). For the LG structure, only the common positions of the β strands are shown. The disulfide bonds are highlighted in yellow. Only β strands (cyan) are labeled for comparison. The α helices of TSPN-1 and CTSP-1 are shown in green. The two structures can’t be simply superimposed. The best alignment can only be made with the middle strands of the back sheet, β14, β5, β10, and β11 of TSPN-1 and β15, β5, β11, and β12 of CTSP-1. Even in this alignment, the β strands from the two front sheets shift over about one strand and also rotate slightly. The edge strands, especially the N-terminal edge strands, are totally out of alignment. The front concaved sheet of CTSP-1 is distinctly curved, while that of TSPN-1 is relatively flat.
Figure 5.
The Sequence Alignment of the N-Terminal Domains of Human TSP-1, TSP-2, TSP-3, and TSP-4; NELL-1 and NELL-2; Collagens α1(XVIII), α1(IX), α1(V), and α1(XI); Xenopus Kielin; and Drosophila TSP
The secondary structures (β strands and α helices) are underlined and are assigned based on the structure of TSPN-1. The irregular, proline-rich strand-like element is marked by a dashed line. The cysteines are colored in yellow and in red shadow. The basic residues contributing to the major heparin binding sites in TSPN-1 are marked in yellow shadow. Those hydrophobic and nonhydrophobic residues that contribute to the core of the TSPN-1 structure or putatively contribute to the cores of other molecule structures are marked in cyan and green shadow, respectively. The RGD triplet in the D-TSP sequence is shadowed in purple. The asparagines that are present within consensus sequences for glycosylation are shown in red. The motif after the last cysteine (e.g., C214 of TSPN-1) in each sequence is colored blue and is assumed to be the linker between a TSPN domain and a helical region that forms either trimeric or pentameric coiled coils. Only part of the linkers of the collagens α1(V), α1(XI), and α1(XVIII) are shown.
The solution of the TSPN-1 structure helps to identify a unique linker (approximately 30 residues) between the globular TSPN-1 domain (N1-C214) and the coiled-coil sequence region that serves as the trimerization site (Figure 5). This model is consistent with electron microscopic images of TSP-1 in which the globular TSPN-1 regions can appear well-separated from each other and from the trimerization site (Lawler et al., 1985). The linker has few predictable secondary structures and is probably very flexible. The sequence and length of the linker vary from molecule to molecule within the group of TSPN-containing proteins. NELLs and Drosophila TSP are exceptional in that they have short linkers. Including the linker in sequence alignments of various TSPNs complicates the analysis of the N-terminal region of these molecules. The TSPN domain is the most N-terminal domain for various proteins, except in a few of the collagens, such as α1(XVIII), etc. (Moradi-Ameli et al., 1994). The linker that follows the TSPN domain generally seems to be flexible and may allow the TSPN domain to adopt different orientations to facilitate ligand binding. In many cases, the TSPN domain has been shown to bind glycosaminoglycans, suggesting that the TSPN domain may serve to anchor the protein to proteoglycans. In this way, the domains in the remainder of the protein are available for other interactions that might direct the cell differentiation, growth, or migration that is associated with the TSPN-containing proteins. The linker may also possess proteolytic cleavage site(s) for the potential release of the TSPN domain as observed in TSP-1 (Damas et al., 2001) and α4(V) collagen (Rothblum et al., 2004). Proteolytic cleavage in the linker region may produce fragments that differ functionally from each other and from the parent molecule. For example, the TSPN-1 domain of TSP-1 stimulates angiogenesis, while the remainder of the protein inhibits angiogenesis (for a review, see Lawler and Detmar, 2004). Cleavage of the linker would also be expected to dissociate the remainder of the molecule from proteoglycans. Thus, proteolysis may represent an important step in the spatial and temporal regulation of functions of the TSPN-containing proteins.
In this study, we have identified a major heparin binding site (including residues R29, K32, R42, R77, K80, K81, and K106) on the bottom of the globular TSPN-1. It has been reported that pairwise mutation of R23 and K24, R28 and R29, or K80 and K81 decreases the affinity of the intact molecule for heparin (Lawler et al., 1992). The localization of R29, K80, and K81 to the positively charged patch on the bottom of the domain is consistent with the mutagenesis data. Whereas R23 and K24 are not included in the major heparin binding site at the bottom of the domain, they are in a flexible loop between the α1 helix and the β2 strand that is in close proximity to it. In this study, we used the pentameric heparin, Arixtra, because it is chemically well-defined and thus more amenable to crystallization. In the structure of the TSPN-1/Arixtra complex, sulfate groups are closely associated with R29, R42, and R77. These data are consistent with the identification of the bottom of TSPN-1 as the heparin binding site and the involvement of R29, as suggested by mutagenesis. The position of R29 shifts significantly in the TSPN-1/Arixtra complex, raising the possibility that glycosaminoglycan binding could affect the functional activity of this portion of the molecule as discussed later. Since R42 is positioned between three sulfate groups, this residue seems to be especially important for Arixtra binding, but its role in other heparin species binding needs further examination. Previous studies have reported that tetrasaccharide heparins can bind TSP-1 and TSPN-1, and that the affinity increases with oligosaccharide length up to decasaccharides (Yu et al., 2000; Mulatero et al., 2003). We hypothesize that the specific interactions involved in heparin binding may vary with the length and sulfation of the oligosaccharide, and that some heparin species interact with K32, K80, K81, and K106, in addition to, or instead of, R29, R42, and R77. We are currently performing crystallization and mutagenesis studies with longer heparins to test this hypothesis.
The three basic residues, R29, R42, and R77, are conserved in TSP-2, suggesting that TSP-1 and -2 may bind heparin through a similar mechanism. Whereas the specific amino acids are not well conserved in other TSPN-containing proteins, homology modeling based on the TSPN-1 structure presented here and the sequence alignment of the proteins shown in Figure 5 reveals the presence of at least one positively charged region in all cases, primarily on the bottom of the domain (data not shown). This is particularly true for Drosophila thrombospondin, which contains five positively charged residues in the region that aligns with R29 and K32 of human TSPN-1. This is consistent with biochemical data showing that many of these proteins bind heparin (Adams et al., 2003; Kuroda et al., 1999; Pihlajamaa et al., 2004).
Besides glycosaminoglycans, a wide variety of protein receptors function to sequester TSP-1 at the cell surface. The binding sites for some of these receptors have been mapped to TSPN-1 by using synthetic peptides. For example, peptide data indicate that calreticulin binds to the region that extends from the α1 helix through the β2 strand (aa 17–35, also designated hep I) (Murphy-Ullrich et al., 1993). This peptide covers the entire α1-β2 loop region (Figure 1A), which is flexible, exposed, and available for interactions. In this sense, the peptide may effectively mimic TSPN-1 in its interaction with calreticulin. Additionally, Arixtra binding, as discussed above, affects the conformation of R29 in the α1-β2 loop. It is possible that cell surface proteoglycans may enhance or inhibit the interaction of TSPN-1 with calreticulin. The interaction with calreticulin may be confined to TSP-1 and -2 because the binding sequence is not present in TSP-3 and -4 or in other related molecules (Figure 5).
The association of TSP-1 with fibrinogen reportedly mediates platelet aggregation and the association of platelets with osteosarcoma cells (Bonnefoy et al., 2001; Voland et al., 2000). Platelet bound fibrinogen reportedly binds to TSP-1 that is on the surface of osteosarcoma cells and may facilitate the hematogenous spread of metastatic cells (Voland et al., 2000). The fibrinogen binding site of TSPN-1 has been mapped with synthetic peptides to a region that includes the β12 strand and the flanking loops (aa 151–164, also designated N12/I) (Voland et al., 2000). This region forms the left edge of the TSPN-1 domain and is solvent accessible (Figure 1A). Since protein-protein interaction interfaces are generally of the size of 1600±400 Å 2 (Lo Conte et al., 1999), the residues from strands β9, β11, and the adjacent loops may also be involved in fibrinogen binding. Additionally, the electrostatic potential is very negative (red in Figure 2A) on the left edge of TSPN-1, suggesting that a positively charged surface patch on fibrinogen may be involved in the interaction.
The proposed fibrinogen binding region partially overlaps with the reported α4β1 integrin binding sequence (A159ELDVP) on the loop between the β12 strand and the α4 helix (Calzada et al., 2004). Synthetic peptide data also indicate that α3β1 and α6β1 bind to TSPN-1 (Calzada et al., 2003; Krutzsch et al., 1999). The α3β1 binding site maps to the β14 strand that runs along the back side of TSPN-1. Of the residues that are reportedly important for binding to α3β1, only R198 is fully exposed on the surface. The α6β1 binding site maps to the β6 strand and the following loop, and Calzada et al. (2003) reported that E90 is essential for α6β1 binding. As shown in Figure 1B, this residue forms salt bridges with R178, R180, and K183 and with a water-mediated hydrogen bond. Thus, the structural data indicate that only the α4β1 site is fully exposed on the surface of TSPN-1. The data presented here will facilitate the design of site-directed mutagenesis strategies to confirm biological activity of the proposed integrin binding sites within the context of the correctly folded domain.
The TSPN-1 structure described here, along with the published structures of the TSRs (Tan et al., 2002), the procollagen homology region (O’Leary et al., 2004), and the last three type 3 repeats with the C-terminal domain (Kvansakul et al., 2004) begin to provide an atomic resolution image of TSP-1. These data permit a detailed comprehension of the structural organization of TSP-1 and its binding sites for proteoglycans, CD36, integrins, and various other secreted and transmembrane proteins. Through these interactions, the TSPs serve their regulatory functions during various forms of tissue remodeling, including synaptogenesis, angiogenesis, wound healing, and neoplasia. Initial studies indicate that the other TSPN-containing proteins may also function during tissue genesis and remodeling.
Experimental Procedures
Preparation of Recombinant TSPN-1
A recombinant version of TSPN-1 (amino acids 1–240 of human TSP-1) was prepared by PCR with the full-length cDNA of human TSP-1 as the template. TSPN-1 was made with the forward primer 573htsp1f (5′-GATGATCCATGGAACCGCATTCCAGAGTCTGGC-3′) and the reverse primer 574htsp1r (5′-GATACCGGTGTTAGTGCGG ATGGCAGGGCT-3′). The PCR product was sequenced and cloned between the NcoI and the AgeI sites of the vector pMT/BiP V5-HisA (Invitrogen; Carlsbad, CA). The recombinant protein includes the vector-derived sequence RSPW at the N terminus and the sequence TGHHHHHH at the C terminus. Vector transfection, cell selection, and protein expression and purification were performed as described previously (Miao et al., 2001). To label the proteins with selenomethionine (Se-Met), the cells were grown to high density (~1 × 107/ml) in media (Hyclone) and were then transferred to methionine-free medium for 4 hr. Se-Met (Sigma) was subsequently added to the medium to a final concentration of 400 mg/l. The cultures were monitored for cell viability and were harvested at 3–5 days. The Se occupancy was estimated to be about 90% based on mass spectral analysis. All proteins were further purified by HPLC in protein buffer of 200 mM NaCl and 20 mM HEPES (pH 7.8).
Crystallization
The purified protein was concentrated to about 10–15 mg/ml for crystallizationwith the vapor diffusion hanging drop method. Protein crystals grew from the buffer containing 30% PEG1500 and 0.08 M NaAc (pH 4.6). Initially, the crystals produced had multiple forms. The majority of them were very thin plates with the space group of P1 (unpublished data). Only one chunky crystal with the space group P21 was obtained. SDS-PAGE of crystal samples indicated that the crystals were actually formed from partially degraded protein. The protein degradation, which was likely caused by unknown residual proteases from the S2 cell expression system, was later detected within days after its concentration. Mass spectral analysis with trypsin-treated, repurified, and concentrated TSPN-1 samples suggested that the degradation was between residues C214 and N230. C214 is present and paired with C153 in the final structure. The loss of the N-linked glycan that is attached to N230 is consistent with the mass reduction and the fact that N230 is the only N-linked glycosylation site in TSPN-1. Limited digestion with α-chymotrypsin (1:100 or 1:400 w/w) was performed with intact TSP-1 for 20 hr at 0°C and was stopped by adding 1 mM PMSF. The principal proteolytic fragment was purified by HPLC or heparin-Sepharose affinity chromatography. The mass spectrum of the α-chymotrypsin-treated protein was very similar to that of the protein formed by the S2 residual proteases. TSPN-1 made by α-chymotrypsin treatment consistently produced thicker P1 crystals. However, the P1 crystals have an unstable lattice and are prone to subtle transformation during heavy atom soaking, posing an obstacle to the structure determination by multiple isomorphous replacement.
It was later found that cocrystallization of TSPN-1 and Arixtra, a synthetic pentameric heparin molecule (Sanofi-Synthelabo, France), consistently produced larger stable crystals of space group P212121 under the same crystallization conditions as that for the unliganded, native forms. The TSPN-1 and Arixtra were mixed in a 1:2 molar ratio prior to crystallization setup. Crystals of the Se-Met-labeled TSPN-1 with Arixtra were also produced under the same crystallization conditions, and they were eventually used for initial phasing.
Data Collection
Diffraction data sets were collected from prefrozen crystals at 100K at the 19ID beamline of the Structure Biology Center at the Advanced Photon Source, Argonne National Laboratory. For structure resolution, the two-wavelength (peak and inflection) inverse-beam MAD (multiwavelength anomalous diffraction) data sets were collected from one Se-Met-labeled TSPN-1/Arixtra cocrystal (Table 1). Another labeled TSPN-1/Arixtra cocrystal was used for acquiring higher-resolution data. Only the peak data set was collected and used for structure refinement of TSPN-1/Arixtra (Table 1). The native data for a crystal of space group P21 were also collected by using the single chunky crystal (Table 1). They were used only for structure refinement. All diffraction data sets were processed and reduced with the HKL2000 suite (Otwinowski and Minor, 1997).
Table 1.
Crystallographic Statistics
| TSPN-1 | TSPN-1/Arixtra |
|||
|---|---|---|---|---|
| Native | Se-Met (1) |
Se-Met (2) |
||
| Peak | Inflection | Peak | ||
| Data Collection | ||||
| Space group | P21 | P212121 | P212121 | P212121 |
| Unit cell | ||||
| a (Å) | 42.812 (1) | 42.345 (1) | 42.391 (1) | 42.404 (1) |
| b (Å) | 41.899 (1) | 53.071 (1) | 53.145 (1) | 53.054 (1) |
| c (Å) | 53.458 (1) | 92.414 (2) | 92.495 (2) | 92.723 (2) |
| α (°) | 90 | 90 | 90 | 90 |
| β (°) | 100.649 (1) | 90 | 90 | 90 |
| γ (°) | 90 | 90 | 90 | 90 |
| Wavelength (Å) | 1.07218 | 0.97923 | 0.97937 | 0.97923 |
| Resolution (Å) | 50–1.8 | 50–2.2 | 50–2.2 | 50–1.9 |
| Number of unique reflections | 17,415 | 19,677a | 19,563a | 30,776a |
| Redundancy | 6.9 | 5.3 | 5.2 | 4.4 |
| Completeness (%) | 99.6 (96.8)b | 96.6 (80.7)b | 93.6 (55.0)b | 96.7 (78.2)b |
| Rmerge (%) | 6.8 (42.7)b | 6.3 (20.8)b | 5.5 (27.3)b | 6.8 (35.9)b |
| I/σ(I) | 32.12 (2.75)b | 29.23 (4.06)b | 30.09 (2.50)b | 27.13 (1.85)b |
| Phasing | ||||
| RCullis (anomalous) (%) | 81 | 84 | ||
| Figure of merit (%) | 64.5 | |||
| Refinement | ||||
| Resolution | 30–1.8 | 30–1.9 | ||
| Reflections (work/test) | 13,196/1,472 | 22,461/2,243 | ||
| Rcrystal/Rfree | 23.64/25.89 | 25.05/27.79 | ||
| Bond length (Å)/angle(°) rms deviation from ideal geometry | 0.007/1.6 | 0.007/1.4 | ||
| Protein atoms average B value (Å2), main chain/side chain | 26.275/27.682 | 33.534/34.576 | ||
Including Bijvoet pairs.
Last resolution bin.
Structure Determination and Refinement
The structure of the TSPN-1/Arixtra complex was determined by two-wavelength MAD phasing (Hendrickson, 1991) of a Se-Met-labeled TSPN-1/Arixtra cocrystal by using the CCP4 suite (CCP4, 1994) (Table 1). Two Se sites were located from anomalous difference Patterson maps, and they were used for initial phasing. After density modification with solvent flattening and histogram mapping, a partial model (including 160 alanines or glycines) was obtained from an automatic model building trial by using the program Resolve (Terwilliger, 2003). It was then examined and reassembled into one molecule through symmetry operations. After sequence fitting and manual model building with the program O (Jones et al., 1991), about 95% of the structure was built. The refinement of the structure was performed with the CNS program suite (Brunger et al., 1998) (Table 1). The final TSPN-1 model contains 206 amino acid residues, from N10 to S215. The only region in the model that does not have electron densities to fit is between residues G185 and V186, and it is located at the tip of a jar handle-like motif as discussed below. The C-terminal residues K213GCS and the residues C153EK on the β9-β10 loop, where the disulfide bond forms, have weak densities. We can’t precisely define the N- and C-terminal residues of the protein because they are not seen in the density maps, and they are likely disordered in the crystals. They are not included in the TSPN-1 structural model. A fraction of Arixtra was built by using the program O. Since O-sulfate and N-sulfate groups of Arixtra are indistinguishable in the structure for the partially disordered molecule, all of them were modeled as O-sulfate groups and designated SO groups.
The native structure of the TSPN-1 in P21 was solved by using the refined TSPN-1 structure obtained from the TSPN-1/Arixtra complex as the search model with the program Molrep in CCP4 suite. The model rebuilding and final refinement of the structure were done with the programs O and CNS, respectively (Table 1). In the final native TSPN-1 model, besides breaking or poor densities observed at the locations mentioned above in the TSPN-1/Arixtra complex, residues A22RKG, which are on a loop between the α1 helix and the β2 strand also have poor densities.
Acknowledgments
We thank Eric Galardi for the expert technical assistance and Lydia Gregg and Sami Lawler for help in preparing the manuscript. This study is supported by National Institutes of Health grants HL68003 and HL49081.
Footnotes
Accession Numbers
The atomic coordinates and structure factors of the native TSPN-1 and the complex TSPN-1/Arixtra have been deposited in the Protein Data Bank with accession codes 1Z78 and 1ZA4, respectively. The atomic coordinates and the structure factors of a higher resolution (1.45 Å) native TSPN-1 have been deposited with access code 2ERF.
References
- Adams JC, Lawler J. The thrombospondins. Int J Biochem Cell Biol. 2004;36:961–968. doi: 10.1016/j.biocel.2004.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams JC, Monk R, Taylor AL, Ozbek S, Fascetti N, Baumgartner S, Engel J. Characterisation of Drosophila thrombospondin defines an early origin of pentameric thrombospondins. J Mol Biol. 2003;328:479–494. doi: 10.1016/s0022-2836(03)00248-1. [DOI] [PubMed] [Google Scholar]
- Bauer KA. New pentasaccharides for prophylaxis of deep vein thrombosis: pharmacology. Chest. 2003;124:364S–370S. doi: 10.1378/chest.124.6_suppl.364s. [DOI] [PubMed] [Google Scholar]
- Beckmann G, Hanke J, Bork P, Reich JG. Merging extracellular domains: fold prediction for laminin G-like and amino-terminal thrombospondin-like modules based on homology to pentraxins. J Mol Biol. 1998;275:725–730. doi: 10.1006/jmbi.1997.1510. [DOI] [PubMed] [Google Scholar]
- Bonnefoy A, Hantgan R, Legrand C, Frojmovic MM. A model of platelet aggregation involving multiple interactions of thrombospondin-1, fibrinogen, and GPIIbIIIa receptor. J Biol Chem. 2001;276:5605–5612. doi: 10.1074/jbc.M010091200. [DOI] [PubMed] [Google Scholar]
- Bork P, Downing AK, Kieffer B, Campbell ID. Structure and distribution of modules in extracellular proteins. Q Rev Biophys. 1996;29:119–167. doi: 10.1017/s0033583500005783. [DOI] [PubMed] [Google Scholar]
- Bornstein P, Sage EH. Matricellular proteins: extracellular modulators of cell function. Curr Opin Cell Biol. 2002;14:608–616. doi: 10.1016/s0955-0674(02)00361-7. [DOI] [PubMed] [Google Scholar]
- Bornstein P, Agah A, Kyriakides TR. The role of thrombospondins 1 and 2 in the regulation of cell-matrix interactions, collagen fibril formation, and the response to injury. Int J Biochem Cell Biol. 2004;36:1115–1125. doi: 10.1016/j.biocel.2004.01.012. [DOI] [PubMed] [Google Scholar]
- Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, et al. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- Calzada MJ, Sipes JM, Krutzsch HC, Yurchenco PD, Annis DS, Mosher DF, Roberts DD. Recognition of the N-terminal modules of thrombospondin-1 and thrombospondin-2 by α6β1 integrin. J Biol Chem. 2003;278:40679–40687. doi: 10.1074/jbc.M302014200. [DOI] [PubMed] [Google Scholar]
- Calzada MJ, Zhou L, Sipes JM, Zhang J, Krutzsch HC, Iruela-Arispe ML, Annis DS, Mosher DF, Roberts DD. α4β1 integrin mediates selective endothelial cell responses to thrombospondins 1 and 2 in vitro and modulates angiogenesis in vivo. Circ Res. 2004;94:462–470. doi: 10.1161/01.RES.0000115555.05668.93. [DOI] [PubMed] [Google Scholar]
- CCP4 (Collaborative Computational Project Number 4) The CCP4 suite: programs for protein crystallography. Acta Crystallogr D Biol Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- Chen H, Herndon ME, Lawler J. The cell biology of thrombospondin-1. Matrix Biol. 2000;19:597–614. doi: 10.1016/s0945-053x(00)00107-4. [DOI] [PubMed] [Google Scholar]
- Christopherson KS, Ullian EM, Stokes CC, Mullowney CE, Hell JW, Agah A, Lawler J, Mosher DF, Bornstein P, Barres BA. Thrombospondins are astrocyte-secreted proteins that promote CNS synaptogenesis. Cell. 2005;120:421–433. doi: 10.1016/j.cell.2004.12.020. [DOI] [PubMed] [Google Scholar]
- Damas C, Vink T, Nieuwenhuis HK, Sixma JJ. The 33-kDa platelet α-granule membrane protein (GMP-33) is an N-terminal proteolytic fragment of thrombospondin. Thromb Haemost. 2001;86:887–893. [PubMed] [Google Scholar]
- Ferrari do Outeiro-Bernstein MA, Nunes SS, Andrade AC, Alves TR, Legrand C, Morandi V. A recombinant NH(2)-terminal heparin-binding domain of the adhesive glycoprotein, thrombospondin-1, promotes endothelial tube formation and cell survival: a possible role for syndecan-4 proteoglycan. Matrix Biol. 2002;21:311–324. doi: 10.1016/s0945-053x(02)00010-0. [DOI] [PubMed] [Google Scholar]
- Hendrickson WA. Determination of macromolecular structures from anomalous diffraction of synchrotron radiation. Science. 1991;254:51–58. doi: 10.1126/science.1925561. [DOI] [PubMed] [Google Scholar]
- Herndon ME, Stipp CS, Lander AD. Interactions of neural glycosaminoglycans and proteoglycans with protein ligands: assessment of selectivity, heterogeneity and the participation of core proteins in binding. Glycobiology. 1999;9:143–155. doi: 10.1093/glycob/9.2.143. [DOI] [PubMed] [Google Scholar]
- Jones TA, Zou JY, Cowan SW, Kjeldgaard Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- Kraulis PJ. Molscript: a program to produce both detailed and schematic plots of proetin structures. J Appl Crystallogr. 1991;24:946–950. [Google Scholar]
- Krutzsch HC, Choe BJ, Sipes JM, Guo N, Roberts DD. Identification of an α(3)β(1) integrin recognition sequence in thrombospondin-1. J Biol Chem. 1999;274:24080–24086. doi: 10.1074/jbc.274.34.24080. [DOI] [PubMed] [Google Scholar]
- Kuroda S, Oyasu M, Kawakami M, Kanayama N, Tanizawa K, Saito N, Abe T, Matsuhashi S, Ting K. Biochemical characterization and expression analysis of neural thrombospondin- 1-like proteins NELL1 and NELL2. Biochem Biophys Res Commun. 1999;265:79–86. doi: 10.1006/bbrc.1999.1638. [DOI] [PubMed] [Google Scholar]
- Kvansakul M, Adams JC, Hohenester E. Structure of a thrombospondin C-terminal fragment reveals a novel calcium core in the type 3 repeats. EMBO J. 2004;23:1223–1233. doi: 10.1038/sj.emboj.7600166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawler J, Detmar M. Tumor progression: the effects of thrombospondin-1 and -2. Int J Biochem Cell Biol. 2004;36:1038–1045. doi: 10.1016/j.biocel.2004.01.008. [DOI] [PubMed] [Google Scholar]
- Lawler J, Derick LH, Connolly JE, Chen JH, Chao FC. The structure of human platelet thrombospondin. J Biol Chem. 1985;260:3762–3772. [PubMed] [Google Scholar]
- Lawler J, Ferro P, Duquette M. Expression and mutagenesis of thrombospondin. Biochemistry. 1992;31:1173–1180. doi: 10.1021/bi00119a029. [DOI] [PubMed] [Google Scholar]
- Lawler J, Sunday M, Thibert V, Duquette M, George EL, Rayburn H, Hynes RO. Thrombospondin-1 is required for normal murine pulmonary homeostasis and its absence causes pneumonia. J Clin Invest. 1998;101:982–992. doi: 10.1172/JCI1684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li SS, Forslow A, Sundqvist KG. Autocrine regulation of T cell motility by calreticulin-thrombospondin-1 interaction. J Immunol. 2005;174:654–661. doi: 10.4049/jimmunol.174.2.654. [DOI] [PubMed] [Google Scholar]
- Lo Conte L, Chothia C, Janin J. The atomic structure of protein-protein recognition sites. J Mol Biol. 1999;285:2177–2198. doi: 10.1006/jmbi.1998.2439. [DOI] [PubMed] [Google Scholar]
- Matsui M, Mizuseki K, Nakatani J, Nakanishi S, Sasai Y. Xenopus kielin: a dorsalizing factor containing multiple chordin-type repeats secreted from the embryonic midline. Proc Natl Acad Sci USA. 2000;97:5291–5296. doi: 10.1073/pnas.090020497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merle B, Malaval L, Lawler J, Delmas P, Clezardin P. Decorin inhibits cell attachment to thrombospondin-1 by binding to a KKTR-dependent cell adhesive site present within the N-terminal domain of thrombospondin-1. J Cell Biochem. 1997;67:75–83. [PubMed] [Google Scholar]
- Miao WM, Seng WL, Duquette M, Lawler P, Laus C, Lawler J. Thrombospondin-1 type 1 repeat recombinant proteins inhibit tumor growth through transforming growth factor-beta-dependent and -independent mechanisms. Cancer Res. 2001;61:7830–7839. [PubMed] [Google Scholar]
- Mikhailenko I, Krylov D, Argraves KM, Roberts DD, Liau G, Strickland DK. Cellular internalization and degradation of thrombospondin-1 is mediated by the amino-terminal heparin binding domain (HBD). High affinity interaction of dimeric HBD with the low density lipoprotein receptor-related protein. J Biol Chem. 1997;272:6784–6791. doi: 10.1074/jbc.272.10.6784. [DOI] [PubMed] [Google Scholar]
- Moradi-Ameli M, Deleage G, Geourjon C, van der Rest M. Common topology within a non-collagenous domain of several different collagen types. Matrix Biol. 1994;14:233–239. doi: 10.1016/0945-053x(94)90187-2. [DOI] [PubMed] [Google Scholar]
- Mulatero C, Lyon M, Lawler J, Jayson G, Gallagher J. Novel approaches to investigate glycoaminoglycan (GAG) binding to the N terminal region of thrombospondin-1 (NTSP) Proc Am Assoc Cancer Res. 2003;44:1430. [Google Scholar]
- Murphy-Ullrich JE, Gurusiddappa S, Frazier WA, Hook M. Heparin-binding peptides from thrombospondins 1 and 2 contain focal adhesion-labilizing activity. J Biol Chem. 1993;268:26784–26789. [PubMed] [Google Scholar]
- Murzin AG, Brenner SE, Hubbard T, Chothia C. SCOP: a structural classification of proteins database for the investigation of sequences and structures. J Mol Biol. 1995;247:536–540. doi: 10.1006/jmbi.1995.0159. [DOI] [PubMed] [Google Scholar]
- Nicholls A, Sharp KA, Honig B. Protein folding and association: insights from the interfacial and thermodynamic properties of hydrocarbons. Proteins. 1991;11:281–296. doi: 10.1002/prot.340110407. [DOI] [PubMed] [Google Scholar]
- O’Leary JM, Hamilton JM, Deane CM, Valeyev NV, Sandell LJ, Downing AK. Solution structure and dynamics of a prototypical chordin-like cysteine-rich repeat (von Willebrand Factor type C module) from collagen IIA. J Biol Chem. 2004;279:53857–53866. doi: 10.1074/jbc.M409225200. [DOI] [PubMed] [Google Scholar]
- Orr AW, Pallero MA, Murphy-Ullrich JE. Thrombospondin stimulates focal adhesion disassembly through Gi- and phosphoinositide 3-kinase-dependent ERK activation. J Biol Chem. 2002;277:20453–20460. doi: 10.1074/jbc.M112091200. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. In: Carter CW Jr, Sweet RM, editors. Methods in Enzymology. New York: Academic Press; 1997. pp. 307–326. [DOI] [PubMed] [Google Scholar]
- Pihlajamaa T, Lankinen H, Ylostalo J, Valmu L, Jaalinoja J, Zaucke F, Spitznagel L, Gosling S, Puustinen A, Morgelin M, et al. Characterization of recombinant amino-terminal NC4 domain of human collagen IX: interaction with glycosaminoglycans and cartilage oligomeric matrix protein. J Biol Chem. 2004;279:24265–24273. doi: 10.1074/jbc.M402865200. [DOI] [PubMed] [Google Scholar]
- Rothblum K, Stahl RC, Carey DJ. Constitutive release of α4 type V collagen N-terminal domain by Schwann cells and binding to cell surface and extracellular matrix heparan sulfate proteoglycans. J Biol Chem. 2004;279:51282–51288. doi: 10.1074/jbc.M408837200. [DOI] [PubMed] [Google Scholar]
- Rudenko G, Hohenester E, Muller YA. LG/LNS domains: multiple functions–one business end? Trends Biochem Sci. 2001;26:363–368. doi: 10.1016/s0968-0004(01)01832-1. [DOI] [PubMed] [Google Scholar]
- Scheel H, Tomiuk S, Hofmann K. A common protein interaction domain links two recently identified epilepsy genes. Hum Mol Genet. 2002;11:1757–1762. doi: 10.1093/hmg/11.15.1757. [DOI] [PubMed] [Google Scholar]
- Tan K, Duquette M, Liu JH, Dong Y, Zhang R, Joachimiak A, Lawler J, Wang JH. Crystal structure of the TSP-1 type 1 repeats: a novel layered fold and its biological implication. J Cell Biol. 2002;159:373–382. doi: 10.1083/jcb.200206062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terwilliger TC. SOLVE and RESOLVE: automated structure solution and density modification. In: Carter CW Jr, Sweet RM, editors. Methods in Enzymology. New York: Academic Press; 2003. pp. 22–37. [DOI] [PubMed] [Google Scholar]
- Voland C, Serre CM, Delmas P, Clezardin P. Platelet-osteosarcoma cell interaction is mediated through a specific fibrinogen-binding sequence located within the N-terminal domain of thrombospondin 1. J Bone Miner Res. 2000;15:361–368. doi: 10.1359/jbmr.2000.15.2.361. [DOI] [PubMed] [Google Scholar]
- Wang S, Herndon ME, Ranganathan S, Godyna S, Lawler J, Argraves WS, Liau G. Internalization but not binding of thrombospondin-1 to low density lipoprotein receptor-related protein-1 requires heparan sulfate proteoglycans. J Cell Biochem. 2004;91:766–776. doi: 10.1002/jcb.10781. [DOI] [PubMed] [Google Scholar]
- Yang Z, Strickland DK, Bornstein P. Extracellular matrix metalloproteinase 2 levels are regulated by the low density lipoprotein-related scavenger receptor and thrombospondin 2. J Biol Chem. 2001;276:8403–8408. doi: 10.1074/jbc.M008925200. [DOI] [PubMed] [Google Scholar]
- Yu H, Tyrrell D, Cashel J, Guo NH, Vogel T, Sipes JM, Lam L, Fillit HM, Hartman J, Mendelovitz S, et al. Specificities of heparin-binding sites from the amino-terminus and type 1 repeats of thrombospondin-1. Arch Biochem Biophys. 2000;374:13–23. doi: 10.1006/abbi.1999.1597. [DOI] [PubMed] [Google Scholar]