Summary
Besides its function as an essential redox cofactor, NAD also serves as consumable substrate for several reactions with broad impact on many cellular processes. NAD homeostasis appears to be tightly controlled, but the mechanism of its regulation is little understood. Here we demonstrate that a previously predicted bacterial transcriptional regulator, NrtR, represses the transcription of NAD biosynthetic genes in vitro. The NAD metabolite ADP-ribose functions as an activator suppressing NrtR repressor activity. The presence of high ADP-ribose levels in the cell is indicative of active NAD turnover in bacteria, which could signal the activation of NAD biosynthetic gene expression via inhibiting the repressor function of NrtR. By comparing the crystal structures of NrtR in complex with DNA and with ADP-ribose, we identified a “Nudix switch” element that likely plays a critical role in the allosteric regulation of DNA binding and repressor function of NrtR.
Introduction
Nicotinamide adenine dinucleotide (NAD) is an indispensable cofactor in all living organisms. In addition to its fundamental function in hundreds of cellular redox reactions, NAD is also a substrate of a number of non-redox enzymes such as bacterial DNA ligase (Wilkinson et al., 2001), various ADP-ribosyltransferases (Holbourn et al., 2006; Koch-Nolte et al., 2008) and protein deacetylases of Sirtuin/CobB family (Denu, 2005; Frye, 2000; Marmorstein, 2004). Depletion of the NAD(P) pool as well as accumulation of certain intermediates, such as quinolinic acid and pyridine mononucleotides, is harmful to the cell. Therefore, biosynthesis and homeostasis of NAD is believed to be a subject of tight regulation. In recent years the NAD biosynthesis machineries consisting of various de novo synthesis, salvage and recycling pathways, has been elucidated in the majority of eukaryotic and prokaryotic species using approaches that combine comparative genomic analysis, metabolic pathways reconstruction, and experimental characterization (Begley et al., 2001; Belenky et al., 2009; Imai, 2009; Magni et al., 2004; Tempel et al., 2007). However, the mechanisms underlying cellular NAD regulation in response to various physiological and environmental conditions are poorly understood. Until recently, two transcriptional factors (TFs) of NAD biosynthesis were known in a limited number of bacterial species: the multifunctional NadR in Enterobacteria acts as a NAD-dependent repressor of de novo and salvage NAD biosynthesis genes (Gerasimova and Gelfand, 2005; Grose et al., 2005; Holley et al., 1985), while YrxA (later renamed NiaR), found in most Bacillus/Clostridium species as well as Fusobacteria and Thermotogales, represses the transcription of certain NAD biosynthetic genes in response to elevated nicotinic acid (NA) levels (Rodionov et al., 2008b; Rossolillo et al., 2005). In an effort to uncover new potential transcriptional regulators of NAD metabolism in bacteria, we applied comparative genomic techniques to analyze NAD biosynthesis subsystems in more than 400 bacterial species and identified a novel TF family, the Nudix-related transcriptional Regulators (NrtR), in a wide range of bacteria lacking NadR and NiaR (Rodionov et al., 2008a). Comparative genomics-based identification of the NrtR-binding DNA motifs (termed NrtR-boxes) allowed in silico reconstruction of NrtR regulons that primarily include genes involved in various aspects of NAD metabolism (Rodionov et al., 2008a). Structural information of two NrtR proteins (pdb codes 2fml and 2fb1) have recently been obtained by Midwest Center for Structural Genomics (MCSG) and revealed that NrtR consists of an N-terminal Nudix hydrolase-like domain (Bessman et al., 1996) and a C-terminal winged-helix-turn-helix (wHTH) domain that is distantly related to histone H5 and several large families of bacterial transcription factors (Aravind et al., 2005; Ramakrishnan et al., 1993). While the wHTH-domain of NrtR is predicted to bind to the operator site of its regulated genes, the Nudix domain is presumably responsible for specific binding of an effector molecule. Among various Nudix hydrolases, ADP-ribose (ADPR) pyrophosphatase (ADPRase), catalyzing ADPR hydrolysis to AMP and ribose 5-phosphate (R5P), shows the most significant similarity to the Nudix domain of NrtR proteins (Rodionov et al., 2008a). Notably, the Nudix domain in most NrtR family members is predicted to be catalytically inactive, as several active site residues in the Nudix-motif and L9 loop regions are no longer conserved (Supplementary Fig. S1). It appears that the ancient Nudix hydrolase domain was recruited by NrtR to function as a signalling module, and might have lost its enzymatic activity but retained the ability to bind (and hence to be regulated by) ADPR, the product of NAD glycohydrolytic reaction.
Here we report the functional and structural characterization of NrtR from a model bacterium Shewanella oneidensis (soNrtR). We demonstrate by an in vitro transcriptional assay that soNrtR is indeed an ADP-ribose dependent transcriptional repressor. The surprisingly high content of ADPR in the cell (0.8 mM, see Result) is indicative of active NAD degradation processes in bacteria, of which very little is currently known. We present the structures of soNrtR in complex with specific NrtR-box DNA and with ligand ADP-ribose. These structures allowed identification of a “Nudix switch” structural element that transduces an ADPR binding signal to the DNA-binding domain of soNrtR.
Results
Organization and transcriptional analysis of the nrtR-nadV-prs region in S. oneidensis genome
The nrtR gene is clustered with genes prs and nadV on the S. oneidensis chromosome, and two NrtR binding sites were predicted in the intergenic region between nrtR and prs (Rodionov et al., 2008a) (Fig. 1A). The prs gene encodes the enzyme ribose phosphate pyrophosphokinase that synthesizes phosphoribosyl pyrophosphate (PRPP), a central precursor for the synthesis of nucleotides (including NAD) and amino acids. The nadV gene encodes the enzyme nicotinamide phosphoribosyltransferase (NamPRT) that catalyzes the transfer of the phosphoribosyl moiety of PRPP to nicotinamide (Nam) forming NMN, the direct NAD precursor. The nadV gene is positioned 10 bp downstream of prs and oriented in the same direction. To investigate whether they form a transcriptional unit, RNA isolated from cells was reverse transcribed and the resulting cDNA was used as a template in PCR with specific primers amplifying the region encompassing prs and part of nadV (region II in Fig. 1A). Amplification of region I in the prs gene was also performed as a positive control. Additional PCR controls utilize genomic DNA and RNA as templates. Amplification of the intergenic region II with cDNA as the template establishes that prs and nadV genes are transcribed as one mRNA molecule (Fig. 1B).
Figure 1. Transcriptional analysis of the nrtR-prs-nadV region.
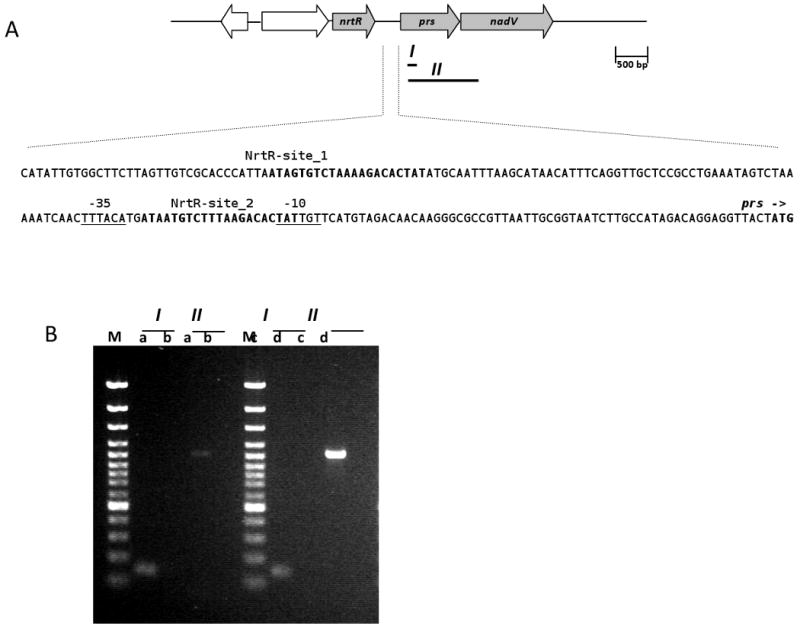
(A) Schematic representation of nrtR, prs and nadV genes on the Shewanella oneidensis chromosome and the promoter region of the prs-nadV operon. I and II represent the PCR-amplified fragments. In the promoter region sequence the two predicted NrtR binding sites are in bold, the putative -10 and -35 regulating sequences are underlined, the start codon of prs is indicated by an arrow and shown in boldface.
(B) Ethidium bromide stained agarose gel showing the PCR products obtained with cDNA (lanes a), RNA (lanes b), genomic DNA (lanes c) and water (lanes d) as the templates and the primer pairs amplifying fragments I and II. Lanes M, 1-kb DNA ladder.
NrtR is a transcriptional repressor
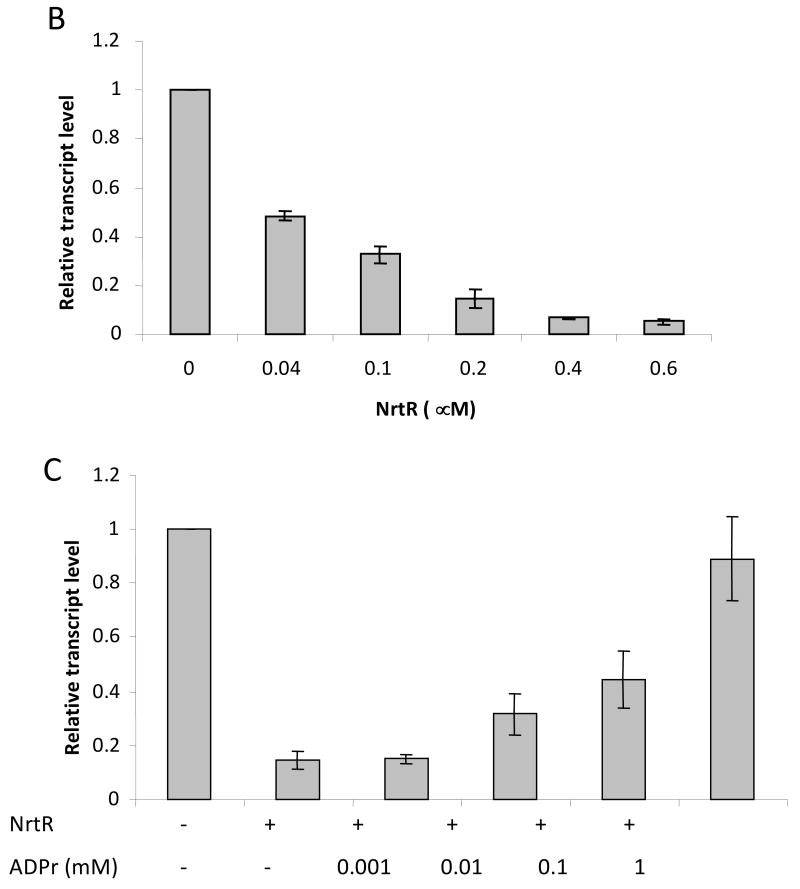
We have previously shown that soNrtR specifically binds to DNA derived from the promoter region of prs-nadV operon containing two predicted NrtR-boxes (Rodionov et al., 2008a). A fluorescence-based DNase I footprinting assay confirmed that the predicted soNrtR-boxes are protected from DNase I digestion in the presence of soNrtR (Fig. 2A). One of the two soNrtR binding sites is located between the predicted promoter elements and partially overlaps the -10 box (Fig. 1A), strongly suggesting that NrtR may act as a transcriptional repressor. To confirm this hypothesis, the effect of NrtR on prs-nadV transcription was tested in vitro with σ70-saturated RNA polymerase from E. coli and a linear DNA template consisting of the promoter region and the 5′ end of the prs gene. The resulting transcript was quantified by reverse transcription (RT)-mediated real-time PCR. As shown in Fig. 2B, transcription was repressed by NrtR in a dose-dependent manner. An unrelated operon was used as a control to verify the specificity of the NrtR effect. The Ct values for the control gene in the absence and presence of 0.2 μM soNrtR were 16.1±0.08 and 15.2 ± 0.03, respectively. These results indicate that soNrtR specifically represses in vitro transcription of the prs-nadV operon.
Figure 2. DNase I footprinting and in vitro transcription assay.
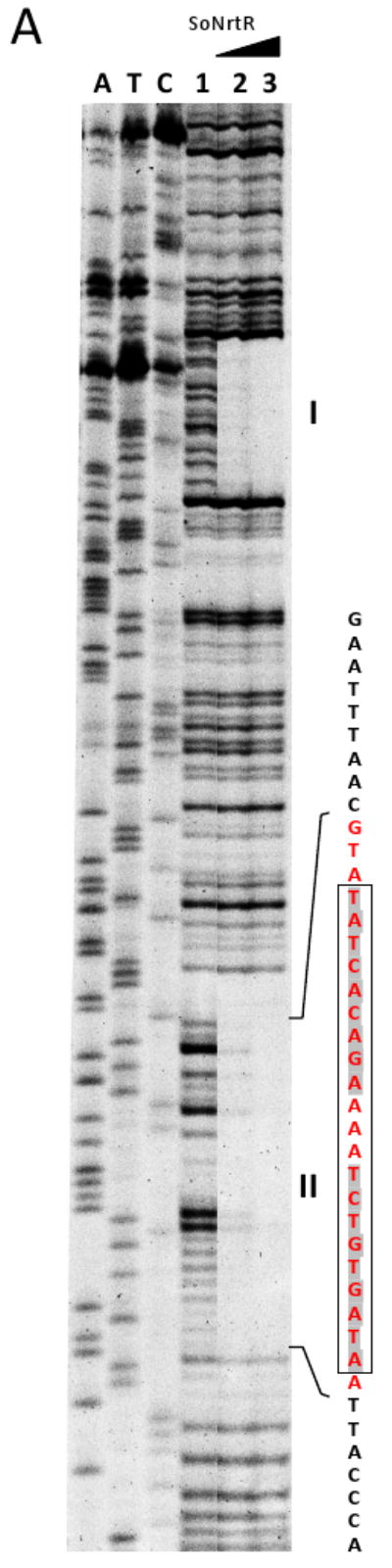
(A) DNase I footprinting of soNrtR on prs-nadV operator DNA. Lanes marked A, T, C are from a DNA sequencing ladder of the coding strand of the 144-bp prs-nadV operator DNA. The remaining lanes show the results of DNase I footprinting. Lane 1 contains no soNrtR; lane 2 and 3 contain increasing amount of soNrtR. The DNA sequence around the second protected region is shown to the right of the gel. Bases protected against DNase I treatment are colored red. The 21-bp NrtR-box sequence is boxed. (B) prs-nadV transcript levels determined by real-time PCR in the presence of various amounts of NrtR. (C) prs-nadV transcript levels in the presence of 0.2 μM NrtrR and increasing amounts of ADPR. Levels of transcripts were normalized to the level in the absence of soNrtR (B) and ADPR (C). For the relative quantification 2 −ΔCt was calculated for each sample (ΔCt being the difference between the Ct values of the target and the normalizer). Data represents the averaged results from three independent experiments with duplicates; vertical bars represent standard deviations.
NAD metabolites (p)ADP-ribose and NaADP are effectors of NrtR
Several evidences, including mobility shift assay data and sequence and structure similarity between the NrtR Nudix domain and ADPRases, suggest that ADPR is the most likely effector molecule of NrtR (Rodionov et al., 2008a). We tested the effect of ADPR on NrtR transcriptional repression by performing the in vitro transcription assay in the presence of various amounts of the metabolite. As shown in Fig. 2C, ADPR was able to reverse the repressive effect of soNrtR in a dose-dependent manner. Repression of transcription was fully abolished in the presence of 1mM ADPR. The same effect was also observed with 1 mM phospho-ADPR (pADPR) and NaADP, while other NAD related metabolites ADP, ribose-5-phosphate, NAD, NADP, NaAD, NADH, NADPH, Nam, NMN and PRPP (at 1 mM concentration) did not show any significant effect on the repressive activity of NrtR (data not shown).
The binding affinities of NAD metabolites to NrtR were determined using a fluorescence-based thermal unfolding assay (Pantoliano et al., 2001). NAD metabolites that do not affect NrtR-DNA interaction and NrtR repressive activity did not significantly affect NrtR thermal stability (not shown). In contrast, protein thermal stability (ΔTm) markedly increased with increasing concentrations of ADPR, pADPR, and NaADP (Fig. 3). The titration profiles resemble saturation kinetic profiles, indicating that these molecules bind specifically. Binding affinities (Kd) of 17.9. 17.7, and 15.4 μM were obtained for ADPR, pADPR and NaADP, respectively (Fig. 3).
Figure 3. Effects of ADPR, pADPR and NaADP on the thermal stability of soNrtR.
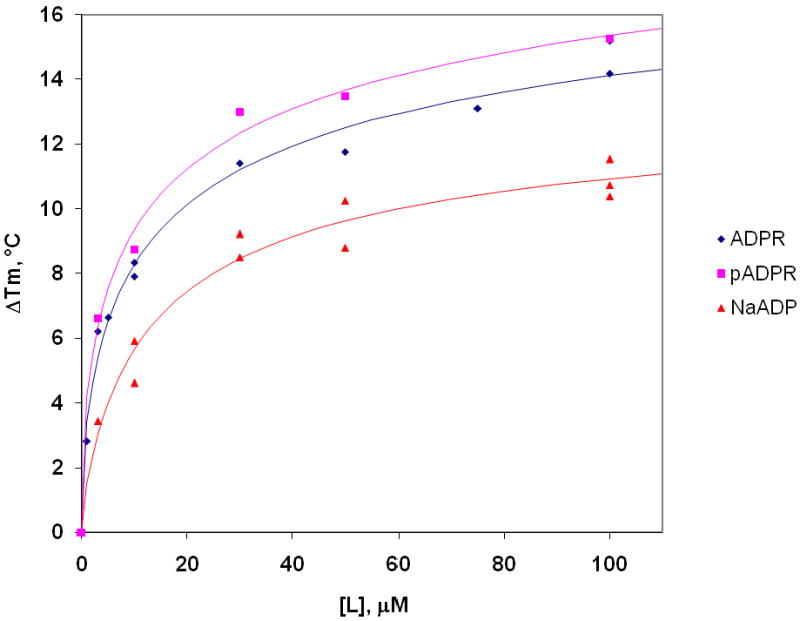
The increase in the thermal stability (ΔTm) of NrtR was monitored in the presence of varying concentrations of ADPR ( ) pADPR (
) pADPR ( ) and NaADP (
) and NaADP ( ).
).
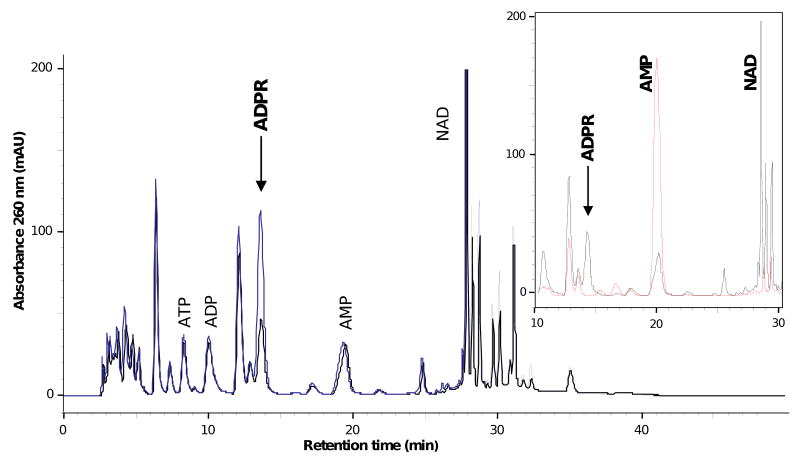
To assess how soNrtR repressor function may be affected by cellular ADPR levels, we determined the concentration of ADPR in S. oneidensis cells. Direct HPLC analysis of cell extracts revealed one single peak (0.97 purity index) that increased in an ADPR spiked sample (Fig. 4). Its identity as genuine ADPR was confirmed by coelution with an ADPR standard (arrow in Fig. 4), identical UV absorption spectra of the sample peak and ADPR standard (not shown), and disappearance of the peak after incubation with nucleotide pyrophosphatase (insert in Fig. 4). From a comparison of the peak areas of the unspiked and spiked samples, a recovery of about 98% during sample preparation was calculated. ADPR in S. oneidensis cells was corrected for recovery and normalized to cell volume, resulting in 1.3 ± 0.2 mM. Control experiments ruled out a possible degradation of NAD to ADPR during sample preparation, but showed that NADH is completely hydrolyzed to ADPR under the experimental condition (not shown). The same observation was also reported by others recently (Tong et al., 2009). Therefore NADH concentration was measured in S. oneidensis cells as described in Methods, resulting in 0.5 ± 0.04 mM. Endogenous ADPR concentration is thus estimated to be 0.8 ± 0.16 mM, after subtracting NADH concentration from the total ADPR. Cellular levels of these nucleotides, as well as of NAD, ATP and ADP are summarized in Supplementary Table S2.
Figure 4. ADPR content in Shewanella oneidensis cells.
HPLC chromatograms of unspiked S. oneidensis extract (black line) and ADPR spiked extract (blue line). Inset shows part of the chromatogram of the unspiked extract (black line) in comparison with that of the same extract after incubation with nucleotide pyrophosphatase (red line)
Overall structure of NrtR
We have determined the structures of apo soNrtR at 2.2Å resolution and in complex with a 28 nucleotide (nt) NrtR-box DNA duplex at 2.9Å resolution. However, repeated attempts to obtain cocrystals of full-length soNrtR in complex with ADPR were not successful. We therefore determined the structure of the N-terminal domain of soNrtR (residue 1-159) in complex with ADPR (referred to as soNrtR_N-ADPR) at 2.4 Å resolution.
The overall structure of soNrtR monomer contains two domains (Fig. 5A). The N-terminal domain displays the Nudix hydrolase-like fold and is most closely related to the ADPRase domain of bacterial NadM-Nudix protein (Huang et al., 2008) with a sequence identity of ∼27% and root mean square deviation (rmsd) between Cα positions of 1.6Å. The soNrtR Nudix domain contains a central mixed five-strand β sheet comprised of β7, β2, β1, β6 and β5, with four α helices (α1- α4) packed on both sides of the sheet (Fig. 5A). The long α4 helix, containing 16 residues, connects the Nudix domain to the C-terminal wHTH DNA binding domain.
Figure 5. Structures of soNrtR, its complexes with DNA, and with effector ADPR.
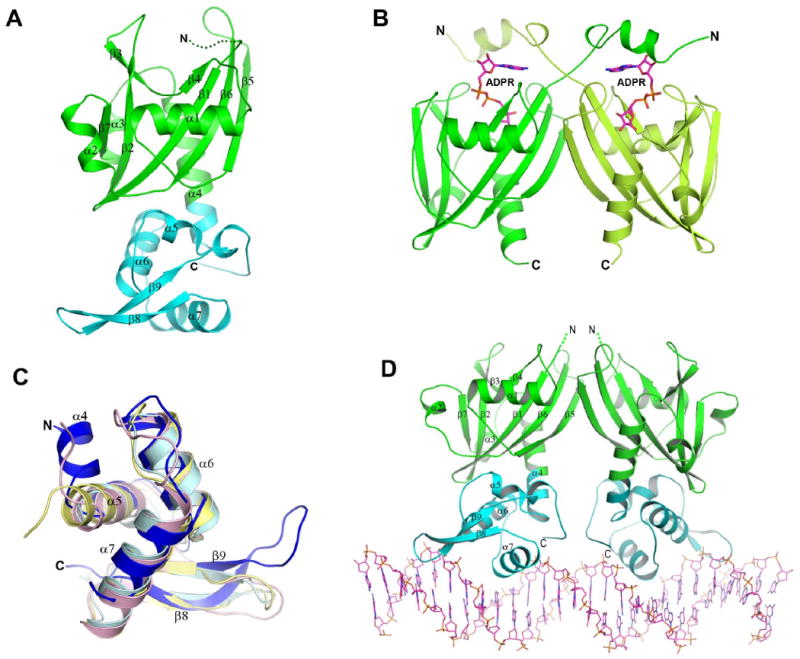
(A). Ribbon presentation of soNrtR monomer with the Nudix domain colored in green and wHTH domain in cyan. The secondary structure elements (strands β1-β9 and helices α1-α7) are labeled. The disordered N-terminal region is indicated by dots.
(B). Structure of soNrtR N-terminal Nudix domain dimer in complex with ADP-ribose. ADP-ribose molecules are shown as magenta sticks.
(C). Superposition of soNrtR wHTH domain (blue) with histone H5 (pink, pdb code 1hst), penicillinase repressor (light cyan, 1sd4), and ferric uptake regulator (yellow, 1mzb).
(D). Structure of full-length soNrtR dimer in complex with DNA. The protein is presented as ribbons and DNA as sticks.
soNrtR forms homodimers in solution (data not shown) and in crystals through Nudix domain interactions (Fig. 5B). Similar to NadM_Nudix proteins (Huang et al., 2008), soNrtR dimerizes mainly through a partial stacking of the central β-sheet from each subunit (Fig. 5B). This dimerization mode is very different from the more distantly related single domain bacterial ADPRases, where dimer interface is composed mostly of a C-terminal α helix (corresponding to α4 of soNrtR) and a N-terminal three-stranded β-sheet domain that is lacking in soNrtR. The N-terminal 18 residues of soNrtR are disordered in the apo and soNrtR-DNA complex structures, but become well ordered upon ADPR binding (Fig. 5B). These residues are structured into a short α helix which is swapped between the two subunits of soNrtR dimer, forming a lid over the bound ADPR (see below for more details).
The DNA-binding wHTH domain of soNrtR consists of a three helical bundle (α5, α6 and α7) followed by a long β-hairpin “wing” (β8 and β9) (Fig. 5C). Structure similarity search using Dali server (Holm and Sander, 1995) identified histone H5 (pdb code 1hst) (Ramakrishnan et al., 1993), penicillinase repressor (1sd4) (Safo et al., 2005), and ferric uptake regulator (1mzb) (Pohl et al., 2003) as the closest structural neighbors. While the relative positioning of secondary structure elements in soNrtR wHTH are similar to these other wHTH domains, there are significant variations in the lengths and conformations of the connecting loops (Fig. 5C). In particular, the β-hairpin wing in soNrtR is much longer than the other wHTH proteins and extended toward a different orientation.
Effector binding site of NrtR
The details of soNrtR ligand binding site are revealed in its complex structure with ADPR (Fig. 6), which are very similar to that observed in NadM_Nudix proteins but different from other bacterial ADPRases (Huang et al., 2008). The adenine ring of ADPR is sandwiched between the side chains of Phe56 from one subunit and Phe15′ from the second subunit of the dimer. The side chain hydroxyl of Tyr10′, also from the second subunit, is hydrogen bonded to both N6 amino and N7 nitrogen groups of the adenine. The positively charged residues Arg41, Lys76 and Arg98 form salt bridges with the oxygen of the diphosphates of ADPR. An extensive hydrogen bonding network exists between the protein and the hydroxyl groups of the ADPR terminal ribose, involving residues Asp23, Arg95, Asp96, and His142 (Fig. 6). Since there is no direct interaction between soNrtR and the adenylyl ribose group, modification at the ribose O2′ position, such as O2′-pADPR, should bind to the protein equally well. This observation is fully consistent with the results from the soNrtR ligand binding assays described in the previous section.
Figure 6. Effector binding site of soNrtR.
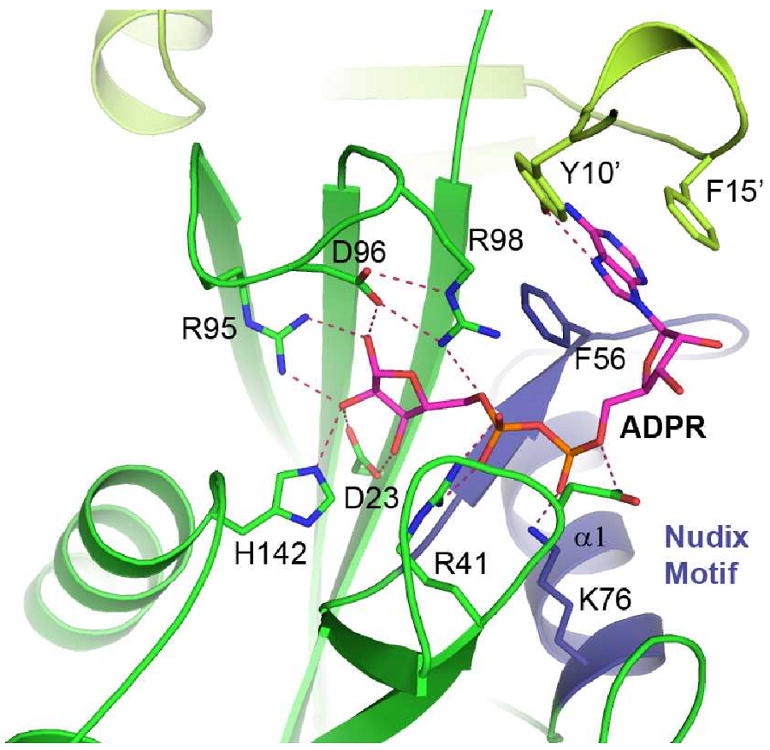
The bound ADPR (magenta) and interacting protein side chains are shown as sticks. Hydrogen bonds are indicated by dashed lines. Residues from the second monomer are colored in pale green and Nudix Motif region is colored blue.
Since the ligand binding assays revealed that NaADP, the product of NADP base-exchange reaction, binds to soNrtR with a similar affinity as ADPR, we modeled a NaADP molecule in the soNrtR ligand binding site (Supplementary Fig. S2). The ADP-ribose moiety was modeled to occupy the same position as the ADPR. The presence of the additional nicotinate base would require that the loop between strand β5 and β6 (residues 92-101) adopt a slightly more open conformation similar to that in the apo soNrtR structure. Residue Phe140 would form a stacking interaction with the pyridine ring and Arg98 may interact with the exocyclic carboxyl group (Supplementary Fig. S2).
NrtR recognizes DNA through its winged-HTH motif
The crystallographic asymmetric unit of the soNrtR-DNA complex contains one soNrtR dimer bound to a 28nt NrtR-box DNA (Fig. 5D). The conformations of the two monomers in the DNA-bound soNrtR dimer are essentially identical (rmsd of 0.22Å for all Cα atoms) and are also very similar to that of apo soNrtR (rmsd 0.61Å), except that the “wing” β-hairpin is better ordered in the complex than in the apo soNrtR structure, apparently through interactions with DNA. In the DNA complex crystal structure, the first 5′- overhanging nucleotide Cyt1 (and Cyt1′) is disordered, while the base-paring 27nt are clearly visible in the electron density (Supplementary Fig. S3). (The prime mark denotes the nucleotide in the complementary strand). The end base pairs, Gua2-Cyt28′ and Gua2′-Cyt28, of the DNA duplex pack against the side chains of Phe46 and Leu49 from adjacent crystallographic symmetry-related soNrtR molecules (Supplementary Fig. S3). These interactions may contribute to a more favorable crystal packing than when shorter nucleotides were used (See Methods).
The 27bp DNA duplex in the complex adopts a slightly distorted B-form conformation (mean rise = 3.3Å, mean twist = 34.5°, and mean roll = 2.6°, mean minor groove width 5.9Å, and mean major groove width 12.5Å). The helical axis of the DNA duplex is mostly straight but slightly bent at both ends, probably due to the inter-crystal lattice stacking interactions with the adjacent soNrtR molecules. Each soNrtR monomer in the dimer interacts with the symmetry related half site of the palindromic NrtR-box DNA in the same manner. Therefore the protein-DNA interactions between one soNrtR monomer and one half site of NrtR-box will be described.
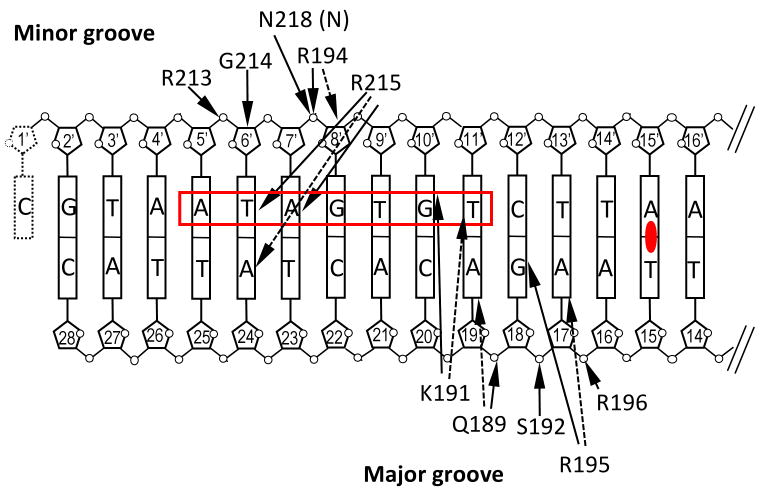
soNrtR interacts with DNA primarily through helix α7, termed “DNA recognition helix”, and the β8-β9 hairpin wing of the wHTH domain. The α7 helix contacts DNA from the major groove side while the β hairpin wing contacts the minor groove (Fig. 7A). The most prominent protein-DNA base interactions are contributed by Arg215 from the tip of the hairpin wing, Lys191 and Arg195 from the DNA recognition helix. The entire side chain of Arg215 is wedged in the minor groove of DNA. Hydrogen bonds are made between the backbone amide of Arg215 to O2 carbonyl of Thy6′ and between the guadininium side chain of Arg215 and N3 of Ade7′. Arg215 is the most conserved residue in NrtR family, where the only other substitution is Lys in a small subset of NrtR proteins (Supplementary Fig. S4). From DNA recognition helix, the side chain of Arg195 forms two hydrogen bonds with Gua18 base (to O6 and N7), and Lys191 side chain is hydrogen bonded to N7 of Gua10′ (Fig. 7A).
Figure 7. Interactions between soNrtR and NrtR-box DNA.
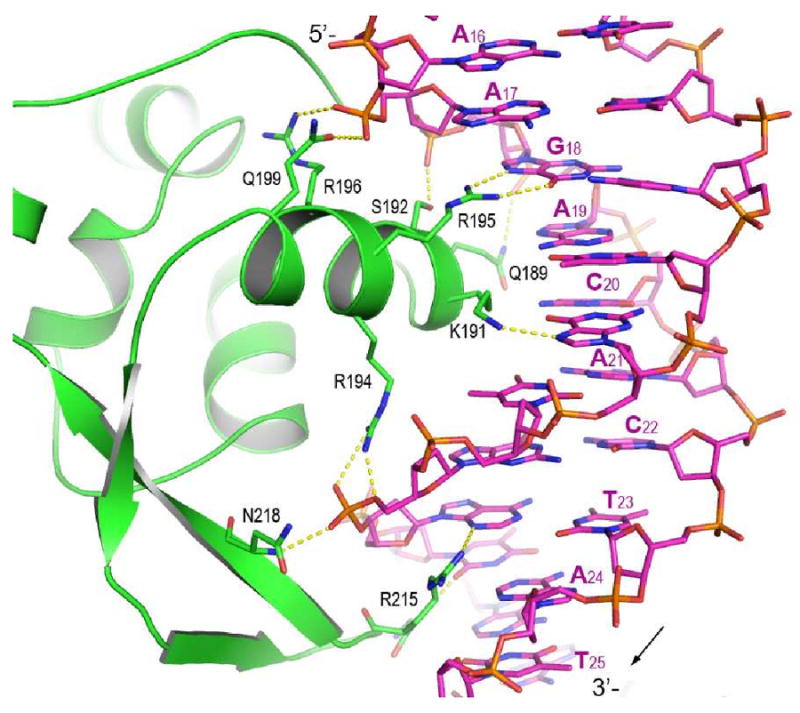
(A). Interactions between soNrtR (green) and one half site of soNrtR-box DNA. Hydrogen bonds are indicated by dotted lines.
(B) Schematic illustration of soNrtR-DNA interactions. The ATA-n2-GT consensus of NrtR-box half site is boxed in red. Solid arrows indicate hydrogen bonds; dashed arrows van der Waals contacts. Primed deoxyribose denotes the complementary DNA strand. The dyad axis of DNA is indicated by a filled oval at the center of base pair Ade15′-Thy15.
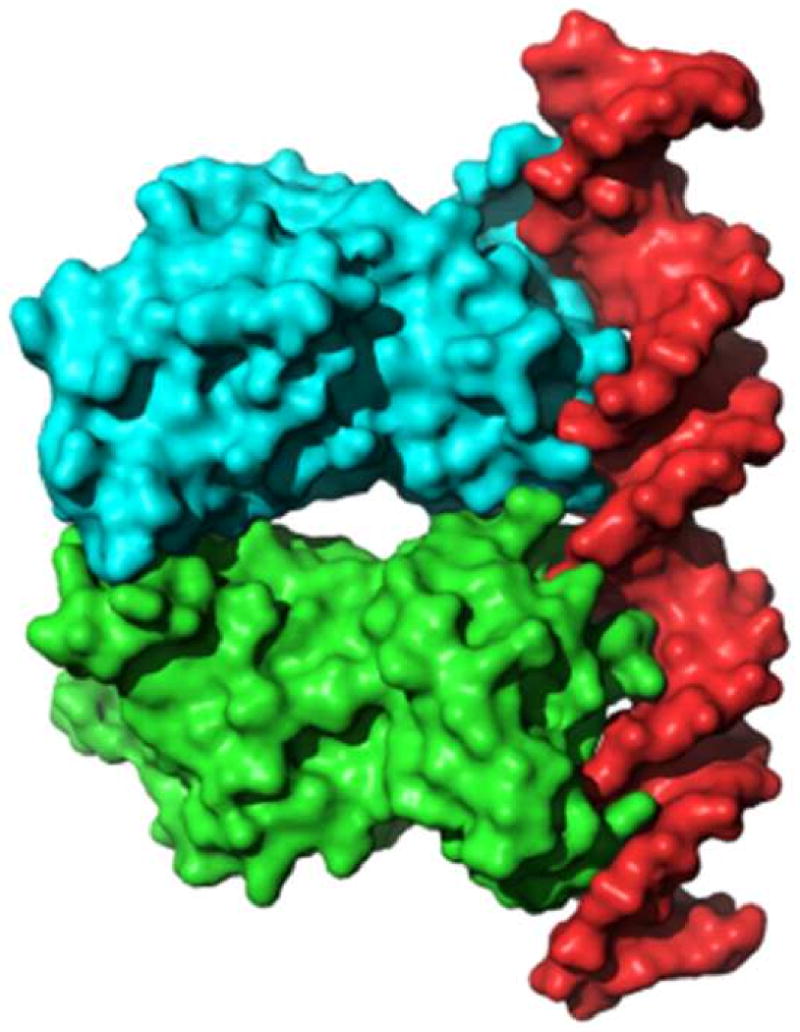
(C). Molecular surface rendering of soNrtR dimer (green and cyan) and DNA duplex (red) in soNrtR-DNA complex.
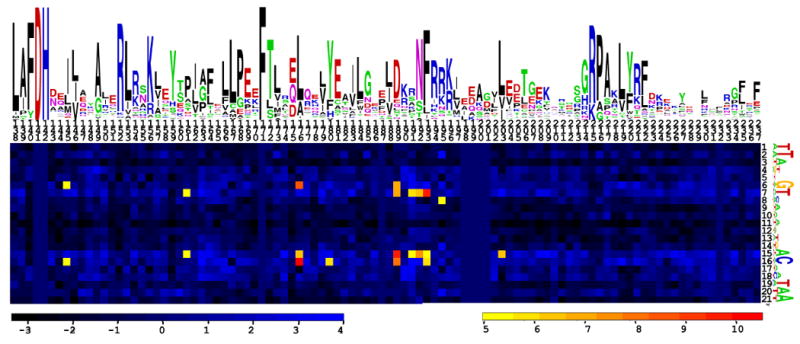
(D). Heatmap of correlations between amino acid-nucleotide pairs for NrtR proteins and their operators. Sequence logos of NrtR DNA-binding domains and NrtR-box DNA are display above and to the right of the heatmap, respectively. The total height of the symbols in each position equals the positional information content, whereas the height of individual symbols is proportional to the positional amino acid/nucleotide frequency. The correlation scores are color ramped from yellow to red for amino acid-nucleotide pairs with statistical significance Z-score > 5.
While there are relatively few sequence specific interactions between soNrtR and NrtR-box DNA, the DNA interacting surface of soNrtR dimer is highly positively charged and several nonspecific polar interactions exist between the protein and DNA backbone phosphates. The side chains of residues Gln189, Ser192, Arg196, Arg194, and Arg213 contact the phosphates of Ade19′, Gua18′, Ade17′, Gua8 and Thy6 nucleotide, respectively (Fig. 7A, 7B). There are also extensive van der Waals interactions between soNrtR and DNA. The protein-DNA interface covers a total of 2392Å2 surface area, about ∼1200 Å2 from each wHTH domain. The shape of this DNA interacting surface, composed from both soNrtR wHTH domains, is alternated with ridges and valleys that complement the major and minor grooves as well as the phosphate backbones of DNA duplex in a largely undistorted B-form conformation (Fig. 7C). The most conserved residue of the NrtR family, Arg215, also has the largest buried surface area (188Å2) upon DNA binding. Other residues with large buried surface upon DNA binding are Arg233 (113Å2), Lys191 (108Å2), Arg195 (92Å2), Gln189 (83Å2), Arg194 (94Å2), and Arg213 (75Å2). Therefore these residues contribute to DNA binding through not only specific hydrogen bonds but also van der Waals interactions.
Co-evolution of NrtR regulators and their DNA recognition motifs
We previously predicted candidate NrtR-binding sites (NrtR-boxes) in most microbial genomes that possess a NrtR transcription factor (Rodionov et al., 2008a). Different NrtR-box consensus sequences were obtained for different NrtR subfamilies on the phylogenetic tree (Supplementary Fig. S5). Significant diversity in NrtR-binding motifs could be correlated with the amino acid sequences of NrtR wHTH domains. Using a multiple sequence alignment of 60 NrtR proteins (Supplementary Fig. S4) and the set of 115 candidate NrtR operators, we computed correlation between amino acid sequences of NrtR wHTH domains and nucleotide sequences of their candidate DNA-binding sites (Fig. 7D). We identified significantly correlated pairs of residues/nucleotides sites (21 pairs total). These sites included ten positions in NrtR proteins and five positions in DNA operators (Supplementary Table S3). Among them, four residues (Gln189, Lys191, Ser192, and Arg195) lie on the DNA recognition helix and make contacts with DNA; five residues (Leu161, Leu176, His180, Phe193, Leu203) form a part of the hydrophobic core of the wHTH domain, and one residue (Leu145) resides on the long α4 helix connecting the wHTH and Nudix domains (see Supplementary Fig. S6).
The first cluster of residues on the DNA recognition helix α7 (Gln189, Lys191, Ser192 and Arg195) may be important for specificity as both Lys191 and Arg195 make base-specific H-bonds with DNA, while Gln189 makes the van der Waals contact with the Ade19 base. In contrast, the most conserved Arg215 has a low correlation score, indicating that it is likely to be important for DNA binding affinity and for recognizing the Ade7 base conserved throughout NrtR family, rather than correlating with subfamily-specific NrtR-box sequences. Interestingly, the second cluster of high scoring correlated residues comprises a part of the hydrophobic core of the wHTH domain (Supplementary Fig. S6). How variations at these positions affect DNA binding specificity is not straightforward to interpret, though they may possibly influence the orientation of the DNA recognition helix α7 and thus specificity. Further experimental investigations of divergent NrtR members are required to achieve a better understanding of their DNA-specificity determinants.
ADP-ribose modulates NrtR repressor function through a Nudix switch
ADPR binding triggers large conformational changes in several regions of soNrtR (Fig. 8). First, the flexible N-terminal 18 residues become well structured upon ADPR binding and swap between the two subunits of the soNrtR dimer so that they interact with the ADPR molecule bound to the adjacent subunit. Aside from the N-terminus, the most drastic conformational changes upon ADPR binding occur in the cross-over linker region connecting strands β6 and β7 (residues 109-125). We term this region “Nudix switch” for reasons discussed below. In apo soNrtR, an α helix (α2) exists in this region, whereas in the ADPR complex structure, the α2 helix completely unwinds and adopts an extended conformation (Fig. 8). Notably, the average B-factors of the α2 helix atoms in the apo soNrtR structure are higher than the rest of the Nudix domain (55 Å2 vs. 43 Å2), suggesting that this region is intrinsically flexible. ADPR binding induced conformational changes appear to stabilize the Nudix switch in the extended conformation as reflected in the better defined density and in the B-factors that are comparable to the rest of the protein (35Å2). This Nudix switch region is equivalent to the flexible “L9 loop” in several other ADPRase and Nudix proteins (Bailey et al., 2002; Gabelli et al., 2001; Gabelli et al., 2002; Huang et al., 2008; Kang et al., 2003), in which two catalytic Asp residues are located at the tip of the loop (Supplementary Fig. S1). Large conformational changes usually occur in this region upon ligand binding to bring the aspartates close to the substrate (Fig. 9). In ADPR bound soNrtR, residue Ser121 at the tip of the loop, corresponding to one of the catalytic Asp of ADPRases, moves ∼16Å toward the ligand binding pocket to interact with ADPR phosphate through a water molecule. Another noted ADPR binding induced conformational change occurs at helix α1, which undergoes an upswing movement (∼2.8Å at the C-terminus of the helix) (Fig. 8). This movement enables the side chain of Lys76 to move about 5.5 Å and directly interact with the diphosphate group of ADPR (Fig. 6). The concerted movements of the α1 helix and Nudix switch region extend the ADPR-binding induced conformational changes to the distal part of the Nudix domain that contacts the DNA-binding wHTH domain in the full-length soNrtR structure. Such a conformational change would result in close contacts (1.8-1.9Å) between the C-terminus of strand β6 (residues 109-111) and helix α5 of the wHTH domain (residues 163-167) (indicated by asterisk in Fig. 8). The wHTH domain would have to adopt a slightly different conformation in the presence of ADPR, which could be detrimental to its DNA binding activity as the precise orientation of the two wHTH domains in soNrtR is important for creating a contiguous interface complementary to the DNA double helix. Additionally, based on the observation that ADPR binding increases susceptibility of the wHTH domain to protease cleavage (data not shown) and that our repeated attempts at obtaining full-length soNrtR-ADPR complex crystals have failed, it is also possible that ADPR induced conformational changes may have a destabilizing effect on the wHTH domain and cause the domain to be more mobile.
Figure 8. ADP-ribose induced conformational changes in soNrtR.
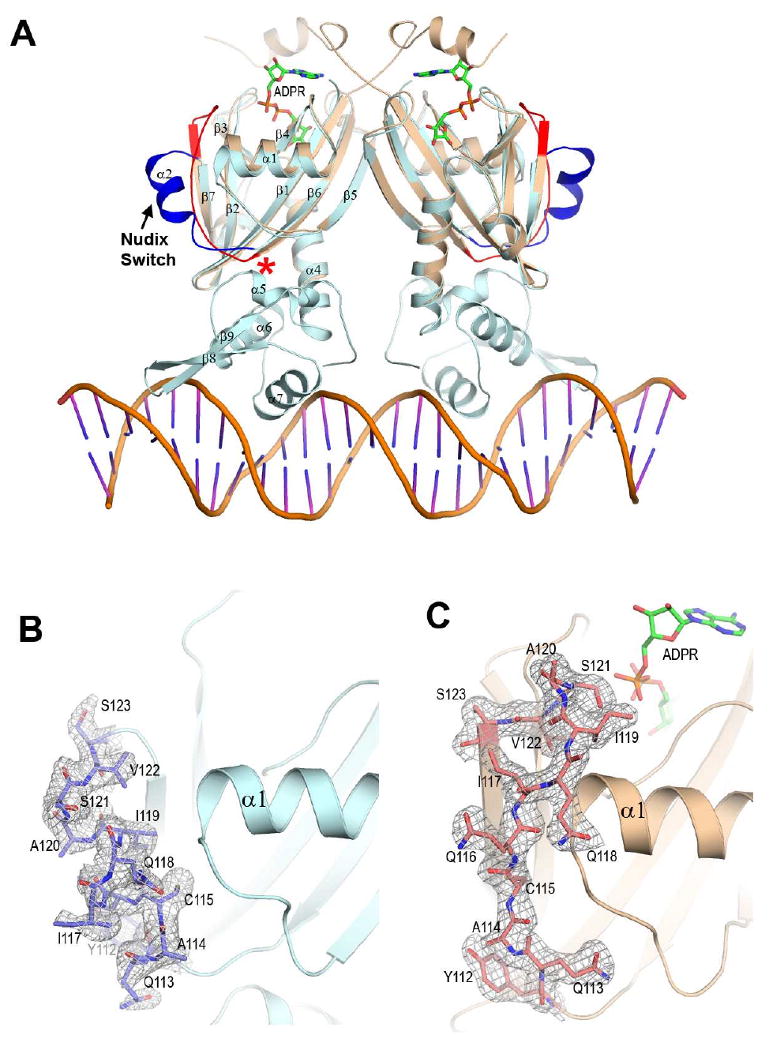
(A). Superposition of soNrtR N-terminal domain dimer in complex with ADPR (colored in wheat) to soNrtR-DNA complex (protein in pale cyan, DNA in orange). The Nudix switch region is indicated by an arrow and highlighted in red and dark blue colors. The region where steric clash would occur is marked by a red asterisk.
(B) and (C). Electron densities of the Nudix Switch region (shown as sticks) in the apo soNrtR (B) and ADPR-bound (C) structures. The densities are contoured at 1.0σ.
Figure 9. Nudix switches in various Nudix proteins.
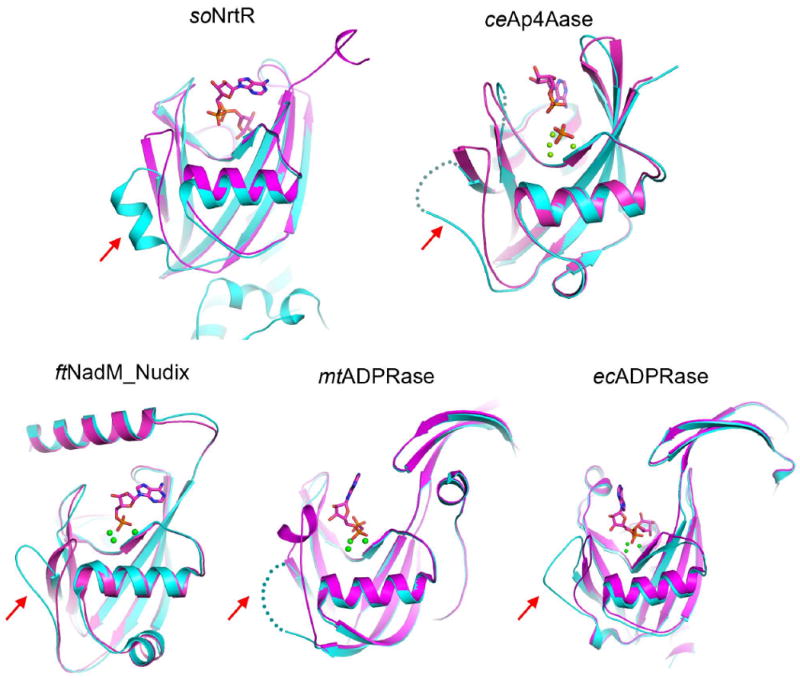
Ligand binding induced conformational changes in following Nudix proteins are shown: soNrtR, C. elegans Ap4A hydrolase (ceAp4Aase), the ADPRase domain of Francisella tularensis NadM_Nudix protein (ftNadM_Nudix), M. tuberculosis ADPRase (mtADPRase), and E. coli ADPRase (ecADPRase). The unliganded (cyan) and liganded (magenta) conformations for each protein are superimposed. The ligands are shown as sticks and metal ions as green balls. The disordered regions in some of the structures are shown as dotted lines. The corresponding Nudix switch region in each structure is indicated by a red arrow.
Discussion
We have previously predicted the existence of a novel family of transcriptional regulators, NrtRs, which may influence various aspects of NAD biosynthesis in many bacterial species (Rodionov et al., 2008a). In the present study, we carried out functional and structural characterization of NrtR from the γ-proteobacterium Shewanella oneidensis, which was predicted to regulate the salvage/recycling of Nam by binding to the promoter region of the operon prs-nadV (Rodionov et al., 2008a). The existence of such an operon would allow a functional coupling of NadV catalyzed NMN synthesis from Nam with the synthesis of PRPP, the cosubstrate of NadV. The subsequent conversion of NMN to NAD would require its deamidation to NaMN by a yet-uncharacterized NMN deamidase before adenylation by the NaMN-specific adenylyltransferase NadD. This interpretation is supported by the experimental verification of a NMN deamidase activity in crude cellular extracts of S. oneidensis (Raffaelli N., unpublished observation).
In the present study we demonstrated that nadV is indeed cotranscribed with prs and that in vitro transcription of this operon is repressed by soNrtR binding to the predicted Nudix-boxes in the promoter region. ADPR, pADPR and NaADP bind NrtR with comparable affinity and abolish the repressor activity. While the physiological relevance of pADPR and NaADP in bacteria remains to be established, ADPR, along with Nam, is known to derive from enzymatic NAD hydrolysis. One possible source of ADPR is the reversible mono-ADP ribosylation of proteins that bacteria use as a mechanism to regulate endogenous protein functions. Well-documented ADP ribosylation targets include dinitrogenase reductase in nitrogen fixing bacteria (Ludden, 1994), RNA polymerase in E. coli cells infected with T4 phage (Goff, 1974), some bacterial glutamine synthetases (Ludden, 1994), membrane proteins involved in nutrients transport in S. coelicolor (Sugawara et al., 2002), and proteins controlling sporulation in B. subtilis (Huh et al., 1996). Additionally, ADPR might be generated from acetyl-ADPR, the product of NAD-dependent protein deacetylases (CobB) of the sirtuin family that are present in various bacteria (Frye, 2000; Zhao et al., 2004). Finally, a mammalian NAD glycohydrolase/ADPR cyclase (CD38) homolog is present in haemolytic streptococci (Kimoto et al., 2006, Michos et al., 2006), and a NAD glycohydrolase activity has been detected in Pseudomonas putida, Bacillus subtilis, and some Mycobacterium species, including M. tuberculosis (Everse et al., 1975; Gopinathan et al., 1964; Mather and Knight, 1972).
In the present work, we report for the first time the ADPR concentration in bacterial cells. In both S. oneidensis (this work) and E. coli (Sorci L. and Osterman A., personal communication), the ADPR levels are in the low mM range, much higher than those determined in human cells, which range from 5 μM to 73 μM, depending on the cell type (Gasser and Guse, 2005; Heiner et al., 2006), and also higher than that in yeast cells (0.1 mM) (Tong et al., 2009). The presence of a consistently high ADPR pool is rather unexpected as free ADP-ribose is considered a highly reactive, potentially toxic molecule that might cause non-enzymatic ADP-ribosylation of proteins (Jacobson et al., 1994). Since cellular ADPR levels are far above the binding constant of the metabolite towards NrtR, it is reasonable to assume that most NrtR proteins in cells are loaded with ADPR and thus do not repress the NAD biosynthetic genes. This assumption is consistent with the view that accumulation of ADPR in the cell is indicative of a very high NAD turnover and may be interpreted as a signal to replenish or maintain active NAD production.
The discovery of a distinct family of ADPR-dependent transcriptional factors revealed a new signalling function of ADPR in transcriptional regulation and further highlights the multifunctionality of this important metabolite. Besides transcriptional regulation, ADPR and related cyclic-ADPR are important secondary messengers for Ca2+ signalling in vertebrate systems (Berger et al., 2004; Lee, 2001; Ying, 2007). In ADPR-gated TRPM2 (or LTRPC2) Ca2+ channels, a cytoplasmic Nudix domain is responsible for ADPR sensing (Kuhn et al., 2005; Perraud et al., 2001). In both cases where ADPR serves as a signalling molecule, the ancient and widespread Nudix hydrolase superfamily protein, in particular the ADPRase, was recruited for ADPR binding and signalling function.
Comparison of the structures of soNrtR-DNA and soNrtR_N-ADPR complexes provided a first glimpse of the structural mechanism underlying the regulatory function of a signaling Nudix domain. The unwinding of helix α2 and associated conformational changes in the Nudix switch region would generate a steric clash that interferes with the proper orientation of the wHTH domain, which may lead to the disruption of protein-DNA interaction. These conformational changes are reminiscent of those observed in several other Nudix proteins (Fig. 9) and suggest that the Nudix switch is a particularly plastic structural element and may serve as a general conformational switch for signaling Nudix proteins.
Another prominent feature of NrtR regulators is their highly varied target DNA sequences in different bacterial species. The structures of soNrtR in complex with its cognate DNA operator reveals that soNrtR dimerizes primarily through interactions between its Nudix domains, and that such dimerization brings the two wHTH DNA-binding domains into proximity to form a contiguous and symmetrical DNA-binding surface that complements the structure of B-form DNA double helix. The binding specificity appears to be determined by a small number of specific interactions between the protein and conserved bases strategically placed along the DNA duplex. Notably, amino acid sequence alignment of NrtR wHTH domains (Supplementary Fig. S4) showed that although the overall sequence conservation of the domain is evident, none of the DNA interacting residues observed from the soNrtR-DNA complex structure are strictly conserved among different NrtR subfamilies. This observation correlates with the highly varied NrtR-box sequences present in different bacterial groups (Supplementary Fig. S5). In the loose consensus of the 21 bp NrtR-box motif, (1)TTAnnGTnnnnnnnACnnTAA(21), soNrtR interacts specifically with the Thy2, Ade3 and Gua6 bases (corresponding to Thy6, Ade7 and Gua10 of the 28-nt DNA used in the soNrtR-DNA complex crystallization. See Fig. 7). A specific protein-base interaction also exists between Arg195 and Gua14 (corresponding to Gua18 in Fig. 7), which is only conserved in a subset of NrtR operators. Statistically significant correlations between NrtR wHTH domain sequences and their DNA operators (Supplementary Table S3) suggest the existence of subfamily-specific protein-DNA interactions, which enables each subfamily of NrtR to specifically recognize its own NrtR-box motif.
Methods
Transcriptional analysis of the nrtR-prs-nadV region
Genomic DNA and RNA were isolated from Shewanella oneidensis MR-1 cells grown in LB medium and collected at an O.D. of 2.0, using the DNeasy Blood & Tissue Kit (Qiagen) and Trizol reagent (Sigma-Aldrich), respectively. Reverse transcription of total RNA was performed in the presence of random primers using the First Strand cDNA Synthesis kit (Biotech. Department Bio Basic Inc.), after DNase I treatment, following kits instructions. PCR was performed on both genomic DNA and reverse transcribed RNA (Supplementary Table S1).
In vitro transcription assay
The DNA template of the target operon (prs-nadV) was generated by using primers prsfw and prsrev (Supplementary Table S1) to amplify the 356-bp promoter region along with the first 440-bp of the prs gene of the S. oneidensis genomic DNA. The transcription reaction mixture contained the DNA template (20 nM), soNrtR and the tested effectors at the indicated concentrations, in 40mM Hepes, pH 7.5, 5 mM DTT, 5 mM MgCl, 2.5% glycerol, 0.5 mg/ml acetylated BSA. After 20 min at room temperature, 20 Units RNase inhibitor and 0.05 Units E. coli σ70-RNA polymerase holoenzyme (Epicentre Technologies) were added and after 15 min at 35°C transcription was initiated by adding 25 μM of each nucleoside triphosphate. Following 15 min incubation at 35°C, the template DNA was digested for 1 hour at 30°C with 2 Units DNase I (Sigma-Aldrich). mRNA transcripts were purified using the SV total RNA isolation system (Promega, Madison, WI, USA) and quantified following reverse transcription by real-time PCR amplifying prs gene internal regions (Supplementary Table S1).
Determination of ADPR concentration in S. oneidensis
S. oneidensis cells were grown in LB medium and collected at the late exponential phase (O.D.600 2.0) by centrifugation (550 × g, 10 min, 4°C). About 4 × 1010 cells were resuspended in 0.66 ml of 100 mM potassium phosphate buffer, pH 6.0 and 0.34 ml of 1.2 M HClO4. After lysis with an ultrasonic disruptor, cell debris was removed by centrifugation (10,000 × g, 10 min, 4°C). The supernatant was divided into two identical samples. To one sample (spiked sample) 8 nmol of ADPR were added. Samples were neutralized with 0.16 M K2CO3, kept on ice for 10 min and centrifuged at 12,000 × g for 3 min. Supernatants were filtered and injected into the HPLC system equipped with a diode-array detector. The enzymatic degradation of ADPR was performed with nucleotide pyrophosphatase, as described in (Heiner et al., 2006). The enzyme was then removed with a Microcon YM10 filter device (Millipore). HPLC separation was performed on a 12.5 cm Supelcosil LC-18-T, 3 μm particle size, reversed-phase column, at 16°C. Elution conditions were 12 min at 100% buffer A (100 mM potassium phosphate buffer, pH 6.0), 8 min up to 12% buffer B (buffer A containing 20% methanol), 3 min up to 45% buffer B, 3 min up to 100% buffer B, holding at 100% buffer B for 9 min and returning to 100% buffer A in 8 min. Flow rate was maintained at 1 ml/min. For NADH determination cells were resuspended in 0.1 M NaOH and after lysis and centrifugation as described above, the supernatant was filtered with a Microcon YM10 filter device (Millipore). NADH concentration was determined in the filtrate spectrophotometrically at 340 nm, with piruvate and lactate dehydrogenase.
Cell number measurement was performed by both direct microscopic counts and plate counts. The average volume of a S. oneidensis cell (0.2 μm3) was calculated based on scanning electron microscope (SEM) images (Hill EA, personal communication).
Crystallization, data collection, and structure determination
Crystallization of soNrtR was carried out using hanging drop vapor diffusion method. 1μl of soNrtR (20 mg/ml) was mixed with equal volume of reservoir solution containing 100 mM sodium citrate pH 6.0 and 2.5 M sodium chloride, and equilibrated against the reservoir at 20 °C until crystal appeared. The crystals were transferred stepwise to a cryoprotection solution containing the original reservoir components and additional 30% glycerol, and frozen in liquid nitrogen.
To obtain heavy atom derivatized soNrtR crystal, quick soaking methods (Dauter et al., 2000) was performed to introduce iodide ion in the crystal. soNrtR crystal was transferred stepwise into a soaking solution containing 1M NaCl, 1 M KI, 100mM sodium citrate pH 6.0 and increasing amount of glycerol up to 30% at 30 seconds each step, and flash frozen in liquid nitrogen.
For the soNrtR-DNA cocrystallization trials, multiple oligodeoxynucleotides containing the NrtR-box sequence with single 5′-overhang and lengths ranging from 25, 26, 27 and 28 nt were used. The best crystals were obtained with the 28nt DNA of sequence 5′-(1)CGTAATAGTGTCTTAAAGACACTATTAC(28) custom synthesized by Sigma-Genosys (nucleotides corresponding to the loose NrtR-box consensus TTAnnGTnnnnnnnACnnTAA are underlined), which was annealed with its complementary strand to form a 27bp duplex with one 5′-Cyt overhang. The protein-DNA complex was formed by mixing soNrtR dimer and annealed DNA duplex at 1:1.5 molar ratio. The final protein concentration is 5mg/ml. The soNrtR-DNA complex crystals were grown at 20°C after mixing equal volume of complex with reservoir solutions containing 0.1 M N-(2-acetamido) iminodiacetic acid (ADA), pH 5.5, 50mM MgCl2 and 8%PEG 4000. The complex crystals were cryoprotected and frozen using a similar procedure as that for apo soNrtR crystals.
The N-terminal domain of soNrtR was generated by chymotrypsin treatment of purified full-length soNrtR in the presence of 200μM ADPR overnight at 4°C. A single band around 15 kDa was observed in the SDS-PAGE after the treatment and mass spectral analysis indicated that the band contained residues 1-159 of soNrtR (denoted as soNrtR_N). soNrtR_N was then purified by Resource Q column (GE Healthcare) and concentrated to 20mg/ml. For cocrystallization with ADPR (final concentration 5mM), 1 μl of soNrtR_N-ADPR complex was mixed with equal volume of the well solution containing 100mM Bis-Tris pH5.5, 20% PEG3350, and 200mM ammonium acetate, and equilibrated against the well for 2-4 days.
All data was collected at X-ray facility at UT Southwestern Medical Center on a Rigaku FR-E rotating anode X-ray generator equipped with RAXIS IV++ image plate detector. The data was processed with HKL2000 (Otwinowski and Minor, 1997). The statistics of all data sets are listed in Table 1. The apo soNrtR structure were determined by single isomorphous replacement/anomalous scattering (SIRAS) phasing method while the N-terminal domain of soNrtR and soNrtR-DNA complex structures were determined by the molecular replacement method (See Supplementary Data).
Table 1. Data collection and refinement statistics.
| apo soNrtR KI derivative | apo soNrtR | soNrtR-DNA complex | soNrtR_N-ADPR complex | |
|---|---|---|---|---|
| Data collection | ||||
| Space group | P3221 | P3221 | P3212 | P212121 |
| Cell dimensions | ||||
| a, b, c (Å) | 75.30, 75.30, 172.53 | 75.16, 75.16, 172.51 | 159.58, 159.58, 73.07 | 63.05, 93.18, 132.50 |
| Resolution (Å) | 2.8 | 2.2 | 2.9 | 2.4 |
| Rsym or Rmerge | 0.080 (0.590)* | 0.039 (0.551) | 0.097 (0.478) | 0.050 (0.274) |
| I / σ(I) | 28.14 (2.49) | 39.17 (2.53) | 20.68 (3.74) | 27.39 (4.35) |
| Completeness (%) | 99.9 (100.0) | 99.6 (99.5) | 98.4 (97.9) | 99.0 (97.6) |
| Redundancy | 4.7 (4.7) | 4.7 (4.6) | 6.1 (6.2) | 4.6 (4.3) |
| Refinement | ||||
| Resolution (Å) | 2.20 | 2.90 | 2.43 | |
| No. reflections | 27872 | 22055 | 28279 | |
| Rwork / Rfree | 0.224/0.279 | 0.200/0.260 | 0.188/0.248 | |
| No. atoms | ||||
| Protein | 3428 | 3544 | 4931 | |
| Ligand/ion | 0 / 2 | 1107 / 1 | 144 / 0 | |
| Water | 216 | 73 | 303 | |
| B-factors (Å2) | ||||
| Protein | 50.96 | 34.22 | 32.46 | |
| Ligand/ion | 66.78 | 42.95 | 32.02 | |
| Water | 53.52 | 29.03 | 34.19 | |
| R.m.s deviations | ||||
| Bond lengths (Å) | 0.013 | 0.014 | 0.014 | |
| Bond angles (°) | 1.59 | 1.87 | 1.79 |
Values in parentheses are for highest-resolution shell.
Supplementary Material
Acknowledgments
Authors wish to thank Drs. Andrei Osterman, Giulio Magni, Nick Grishin and Silverio Ruggieri for helpful discussion; Dr. Lisa Kinch for critical reading of the manuscript, Dr. Adolfo Amici for technical assistance; Dr. Eric Allen Hill for providing the S. oneidensis cell volume information. This work was supported by a Welch Foundation grant (I-5015) and in part by NIH grant AI059146. The work of D. A. R. was partially supported by the grant “Integrated Biology of Shewanella” (PI J. Fredrikson) from the U. S. Department of Energy, Office of biological and Environmental Research under the Genomics:GTL Program. M.S.G., A.B.R. and Y.D.K. were partially supported by grants from the Howard Hughes Medical Institute (55005610) and the program “Molecular and Cellular Biology” of the Russian Academy of Sciences to M.S.G.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errorsmaybe discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Accession codes Atomic coordinates of apo soNrtR, soNrtR-DNA and soNrtR_N-ADPR complexes have been deposited in the Protein Data Bank with accession codes 3GZ5, 3GZ6, and 3GZ8.
References
- Aravind L, Anantharaman V, Balaji S, Babu MM, Iyer LM. The many faces of the helix-turn-helix domain: transcription regulation and beyond. FEMS Microbiol Rev. 2005;29:231–262. doi: 10.1016/j.femsre.2004.12.008. [DOI] [PubMed] [Google Scholar]
- Bailey S, Sedelnikova SE, Blackburn GM, Abdelghany HM, Baker PJ, McLennan AG, Rafferty JB. The crystal structure of diadenosine tetraphosphate hydrolase from Caenorhabditis elegans in free and binary complex forms. Structure. 2002;10:589–600. doi: 10.1016/s0969-2126(02)00746-3. [DOI] [PubMed] [Google Scholar]
- Begley TP, Kinsland C, Mehl RA, Osterman A, Dorrestein P. The biosynthesis of nicotinamide adenine dinucleotides in bacteria. Vitam Horm. 2001;61:103–119. doi: 10.1016/s0083-6729(01)61003-3. [DOI] [PubMed] [Google Scholar]
- Belenky P, Christensen KC, Gazzaniga F, Pletnev AA, Brenner C. Nicotinamide riboside and nicotinic acid riboside salvage in fungi and mammals. Quantitative basis for Urh1 and purine nucleoside phosphorylase function in NAD+ metabolism. J Biol Chem. 2009;284:158–164. doi: 10.1074/jbc.M807976200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger F, Ramirez-Hernandez MH, Ziegler M. The new life of a centenarian: signalling functions of NAD(P) Trends Biochem Sci. 2004;29:111–118. doi: 10.1016/j.tibs.2004.01.007. [DOI] [PubMed] [Google Scholar]
- Bessman MJ, Frick DN, O'Handley SF. The MutT proteins or “Nudix” hydrolases, a family of versatile, widely distributed, “housecleaning” enzymes. J Biol Chem. 1996;271:25059–25062. doi: 10.1074/jbc.271.41.25059. [DOI] [PubMed] [Google Scholar]
- Dauter Z, Dauter M, Rajashankar KR. Novel approach to phasing proteins: derivatization by short cryo-soaking with halides. Acta Crystallogr D Biol Crystallogr. 2000;56:232–237. doi: 10.1107/s0907444999016352. [DOI] [PubMed] [Google Scholar]
- Denu JM. The Sir 2 family of protein deacetylases. Curr Opin Chem Biol. 2005;9:431–440. doi: 10.1016/j.cbpa.2005.08.010. [DOI] [PubMed] [Google Scholar]
- Everse J, Everse KE, Kaplan NO. The pyridine nucleosidases from Bacillus subtilis and Neurospora crassa. Isolation and structural properties. Arch Biochem Biophys. 1975;169:702–713. doi: 10.1016/0003-9861(75)90215-5. [DOI] [PubMed] [Google Scholar]
- Frye RA. Phylogenetic classification of prokaryotic and eukaryotic Sir2-like proteins. Biochem Biophys Res Commun. 2000;273:793–798. doi: 10.1006/bbrc.2000.3000. [DOI] [PubMed] [Google Scholar]
- Gabelli SB, Bianchet MA, Bessman MJ, Amzel LM. The structure of ADP-ribose pyrophosphatase reveals the structural basis for the versatility of the Nudix family. Nat Struct Biol. 2001;8:467–472. doi: 10.1038/87647. [DOI] [PubMed] [Google Scholar]
- Gabelli SB, Bianchet MA, Ohnishi Y, Ichikawa Y, Bessman MJ, Amzel LM. Mechanism of the Escherichia coli ADP-ribose pyrophosphatase, a Nudix hydrolase. Biochemistry. 2002;41:9279–9285. doi: 10.1021/bi0259296. [DOI] [PubMed] [Google Scholar]
- Gasser A, Guse AH. Determination of intracellular concentrations of the TRPM2 agonist ADP-ribose by reversed-phase HPLC. J Chromatogr B Analyt Technol Biomed Life Sci. 2005;821:181–187. doi: 10.1016/j.jchromb.2005.05.002. [DOI] [PubMed] [Google Scholar]
- Gerasimova AV, Gelfand MS. Evolution of the NadR regulon in Enterobacteriaceae. J Bioinform Comput Biol. 2005;3:1007–1019. doi: 10.1142/s0219720005001387. [DOI] [PubMed] [Google Scholar]
- Goff CG. Chemical structure of a modification of the Escherichia coli ribonucleic acid polymerase alpha polypeptides induced by bacteriophage T4 infection. J Biol Chem. 1974;249:6181–6190. [PubMed] [Google Scholar]
- Gopinathan KP, Sirsi M, Vaidyanathan CS. Nicotinamide-adenine dinucleotide glycohydrolase of Mycobacterium tuberculosis H37Rv. Biochem J. 1964;91:277–282. doi: 10.1042/bj0910277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grose JH, Bergthorsson U, Roth JR. Regulation of NAD synthesis by the trifunctional NadR protein of Salmonella enterica. J Bacteriol. 2005;187:2774–2782. doi: 10.1128/JB.187.8.2774-2782.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heiner I, Eisfeld J, Warnstedt M, Radukina N, Jungling E, Luckhoff A. Endogenous ADP-ribose enables calcium-regulated cation currents through TRPM2 channels in neutrophil granulocytes. Biochem J. 2006;398:225–232. doi: 10.1042/BJ20060183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holbourn KP, Shone CC, Acharya KR. A family of killer toxins. Exploring the mechanism of ADP-ribosylating toxins. Febs J. 2006;273:4579–4593. doi: 10.1111/j.1742-4658.2006.05442.x. [DOI] [PubMed] [Google Scholar]
- Holley EA, Spector MP, Foster JW. Regulation of NAD biosynthesis in Salmonella typhimurium: expression of nad-lac gene fusions and identification of a nad regulatory locus. J Gen Microbiol. 1985;131:2759–2770. doi: 10.1099/00221287-131-10-2759. [DOI] [PubMed] [Google Scholar]
- Holm L, Sander C. Dali: a network tool for protein structure comparison. Trends Biochem Sci. 1995;20:478–480. doi: 10.1016/s0968-0004(00)89105-7. [DOI] [PubMed] [Google Scholar]
- Huang N, Sorci L, Zhang X, Brautigam CA, Li X, Raffaelli N, Magni G, Grishin NV, Osterman AL, Zhang H. Bifunctional NMN adenylyltransferase/ADP-ribose pyrophosphatase: structure and function in bacterial NAD metabolism. Structure. 2008;16:196–209. doi: 10.1016/j.str.2007.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huh JW, Shima J, Ochi K. ADP-ribosylation of proteins in Bacillus subtilis and its possible importance in sporulation. J Bacteriol. 1996;178:4935–4941. doi: 10.1128/jb.178.16.4935-4941.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai S. Nicotinamide phosphoribosyltransferase (Nampt): a link between NAD biology, metabolism, and diseases. Curr Pharm Des. 2009;15:20–28. doi: 10.2174/138161209787185814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobson EL, Cervantes-Laurean D, Jacobson MK. Glycation of proteins by ADP-ribose. Mol Cell Biochem. 1994;138:207–212. doi: 10.1007/BF00928463. [DOI] [PubMed] [Google Scholar]
- Kang LW, Gabelli SB, Cunningham JE, O'Handley SF, Amzel LM. Structure and mechanism of MT-ADPRase, a nudix hydrolase from Mycobacterium tuberculosis. Structure. 2003;11:1015–1023. doi: 10.1016/s0969-2126(03)00154-0. [DOI] [PubMed] [Google Scholar]
- Kimoto H, Fujii Y, Hirano S, Yokota Y, Taketo A. Genetic and biochemical properties of streptococcal NAD-glycohydrolase inhibitor. J Biol Chem. 2006;281:9181–9189. doi: 10.1074/jbc.M506879200. [DOI] [PubMed] [Google Scholar]
- Koch-Nolte F, Kernstock S, Mueller-Dieckmann C, Weiss MS, Haag F. Mammalian ADP-ribosyltransferases and ADP-ribosylhydrolases. Front Biosci. 2008;13:6716–6729. doi: 10.2741/3184. [DOI] [PubMed] [Google Scholar]
- Kuhn FJ, Heiner I, Luckhoff A. TRPM2: a calcium influx pathway regulated by oxidative stress and the novel second messenger ADP-ribose. Pflugers Arch. 2005;451:212–219. doi: 10.1007/s00424-005-1446-y. [DOI] [PubMed] [Google Scholar]
- Lee HC. Physiological functions of cyclic ADP-ribose and NAADP as calcium messengers. Annu Rev Pharmacol Toxicol. 2001;41:317–345. doi: 10.1146/annurev.pharmtox.41.1.317. [DOI] [PubMed] [Google Scholar]
- Ludden PW. Reversible ADP-ribosylation as a mechanism of enzyme regulation in procaryotes. Mol Cell Biochem. 1994;138:123–129. doi: 10.1007/BF00928453. [DOI] [PubMed] [Google Scholar]
- Magni G, Amici A, Emanuelli M, Orsomando G, Raffaelli N, Ruggieri S. Enzymology of NAD+ homeostasis in man. Cell Mol Life Sci. 2004;61:19–34. doi: 10.1007/s00018-003-3161-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marmorstein R. Structure and chemistry of the Sir2 family of NAD+-dependent histone/protein deactylases. Biochem Soc Trans. 2004;32:904–909. doi: 10.1042/BST0320904. [DOI] [PubMed] [Google Scholar]
- Mather LH, Knight M. A heat-stable nicotinamide-adenine dinucleotide glycohydrolase from Pseudomonas putida KB1. Partial purification and some properties of the enzyme and an inhibitory protein. Biochem J. 1972;129:141–152. doi: 10.1042/bj1290141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michos A, Gryllos I, Hakansson A, Srivastava A, Kokkotou E, Wessels MR. Enhancement of streptolysin O activity and intrinsic cytotoxic effects of the group A streptococcal toxin, NAD-glycohydrolase. J Biol Chem. 2006;281:8216–8223. doi: 10.1074/jbc.M511674200. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- Pantoliano MW, Petrella EC, Kwasnoski JD, Lobanov VS, Myslik J, Graf E, Carver T, Asel E, Springer BA, Lane P, Salemme FR. High-density miniaturized thermal shift assays as a general strategy for drug discovery. J Biomol Screen. 2001;6:429–440. doi: 10.1177/108705710100600609. [DOI] [PubMed] [Google Scholar]
- Perraud AL, Fleig A, Dunn CA, Bagley LA, Launay P, Schmitz C, Stokes AJ, Zhu Q, Bessman MJ, Penner R, et al. ADP-ribose gating of the calcium-permeable LTRPC2 channel revealed by Nudix motif homology. Nature. 2001;411:595–599. doi: 10.1038/35079100. [DOI] [PubMed] [Google Scholar]
- Pohl E, Haller JC, Mijovilovich A, Meyer-Klaucke W, Garman E, Vasil ML. Architecture of a protein central to iron homeostasis: crystal structure and spectroscopic analysis of the ferric uptake regulator. Mol Microbiol. 2003;47:903–915. doi: 10.1046/j.1365-2958.2003.03337.x. [DOI] [PubMed] [Google Scholar]
- Ramakrishnan V, Finch JT, Graziano V, Lee PL, Sweet RM. Crystal structure of globular domain of histone H5 and its implications for nucleosome binding. Nature. 1993;362:219–223. doi: 10.1038/362219a0. [DOI] [PubMed] [Google Scholar]
- Rodionov DA, De Ingeniis J, Mancini C, Cimadamore F, Zhang H, Osterman AL, Raffaelli N. Transcriptional regulation of NAD metabolism in bacteria: NrtR family of Nudix-related regulators. Nucleic Acids Res. 2008a;36:2047–2059. doi: 10.1093/nar/gkn047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodionov DA, Li X, Rodionova IA, Yang C, Sorci L, Dervyn E, Martynowski D, Zhang H, Gelfand MS, Osterman AL. Transcriptional regulation of NAD metabolism in bacteria: genomic reconstruction of NiaR (YrxA) regulon. Nucleic Acids Res. 2008b;36:2032–2046. doi: 10.1093/nar/gkn046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossolillo P, Marinoni I, Galli E, Colosimo A, Albertini AM. YrxA is the transcriptional regulator that represses de novo NAD biosynthesis in Bacillus subtilis. J Bacteriol. 2005;187:7155–7160. doi: 10.1128/JB.187.20.7155-7160.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Safo MK, Zhao Q, Ko TP, Musayev FN, Robinson H, Scarsdale N, Wang AH, Archer GL. Crystal structures of the BlaI repressor from Staphylococcus aureus and its complex with DNA: insights into transcriptional regulation of the bla and mec operons. J Bacteriol. 2005;187:1833–1844. doi: 10.1128/JB.187.5.1833-1844.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugawara K, Dohmae N, Kasai K, Saido-Sakanaka H, Okamoto S, Takio K, Ochi K. Isolation and identification of novel ADP-ribosylated proteins from Streptomyces coelicolor A3(2) Biosci Biotechnol Biochem. 2002;66:2292–2296. doi: 10.1271/bbb.66.2292. [DOI] [PubMed] [Google Scholar]
- Tempel W, Rabeh WM, Bogan KL, Belenky P, Wojcik M, Seidle HF, Nedyalkova L, Yang T, Sauve AA, Park HW, Brenner C. Nicotinamide riboside kinase structures reveal new pathways to NAD+ PLoS Biol. 2007;5:e263. doi: 10.1371/journal.pbio.0050263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong L, Lee S, Denu JM. Hydrolase regulates NAD metabolites and modulates cellular REDOX. J Biol Chem. 2009 doi: 10.1074/jbc.M809790200. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson A, Day J, Bowater R. Bacterial DNA ligases. Mol Microbiol. 2001;40:1241–1248. doi: 10.1046/j.1365-2958.2001.02479.x. [DOI] [PubMed] [Google Scholar]
- Ying W. NAD+ and NADH in brain functions, brain diseases and brain aging. Front Biosci. 2007;12:1863–1888. doi: 10.2741/2194. [DOI] [PubMed] [Google Scholar]
- Zhao K, Chai X, Marmorstein R. Structure and substrate binding properties of cobB, a Sir2 homolog protein deacetylase from Escherichia coli. J Mol Biol. 2004;337:731–741. doi: 10.1016/j.jmb.2004.01.060. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.