Abstract
DNA methylation is essential for normal development1–3 and has been implicated in many pathologies including cancer4,5. Our knowledge about the genome-wide distribution of DNA methylation, how it changes during cellular differentiation and how it relates to histone methylation and other chromatin modifications in mammals remains limited. Here we report the generation and analysis of genome-scale DNA methylation profiles at nucleotide resolution in mammalian cells. Using high-throughput reduced representation bisulphite sequencing6 and single-molecule-based sequencing, we generated DNA methylation maps covering most CpG islands, and a representative sampling of conserved non-coding elements, transposons and other genomic features, for mouse embryonic stem cells, embryonic-stem-cell-derived and primary neural cells, and eight other primary tissues. Several key findings emerge from the data. First, DNA methylation patterns are better correlated with histone methylation patterns than with the underlying genome sequence context. Second, methylation of CpGs are dynamic epigenetic marks that undergo extensive changes during cellular differentiation, particularly in regulatory regions outside of core promoters. Third, analysis of embryonic-stem-cell-derived and primary cells reveals that ‘weak’ CpG islands associated with a specific set of developmentally regulated genes undergo aberrant hypermethylation during extended proliferation in vitro, in a pattern reminiscent of that reported in some primary tumours. More generally, the results establish reduced representation bisulphite sequencing as a powerful technology for epigenetic profiling of cell populations relevant to developmental biology, cancer and regenerative medicine.
DNA methylation can be detected by sequencing genomic DNA that has been treated with sodium bisulphite7. It has been impractical to apply bisulphite sequencing at a genome-wide scale because polymerase chain reaction (PCR)-based8 and whole-genome shotgun9 approaches are currently too inefficient for comparative analysis across multiple cell states in large mammalian genomes. However, reduced representations can be generated to sequence a defined fraction of a large genome6,10. Computational analysis indicated that digesting mouse genomic DNA with the methylation-insensitive restriction enzyme MspI, selecting 40–220-base pair (bp) fragments, and performing 36-bp end-sequencing would cover ∼1 million distinct CpG dinucleotides (4.8% of all CpGs), with roughly half located within ‘CpG islands’ (including sequences from 90% of all CpG islands) and the rest distributed between other relatively CpG-poor sequence features (Supplementary Fig. 1 and Supplementary Table 1). Notably, although CpGs are not distributed uniformly in the genome, every MspI reduced representation bisulphite sequencing (RRBS) sequence read includes at least one informative CpG position (Supplementary Fig. 2), making the approach highly efficient.
We validated high-throughout RRBS by sequencing MspI fragments from wild-type and methylation-deficient embryonic stem (ES) cells6, using an Illumina Genome Analyser. We generated an initial set of ∼21 million high quality, aligned RRBS reads. The reads from each cell type included ∼97% of the predicted non-repetitive MspI fragments (12-fold and 8-fold median coverage, respectively). This demonstrates that RRBS library construction is relatively unbiased (Supplementary Fig. 3) and is insensitive to genome-wide CpG methylation levels (estimated by nearest-neighbour analysis as 72% and 0.5%, respectively). Reads from both cell types showed near complete (>99%) bisulphite conversion of non-CpG cytosines.
To investigate cell-type-specific DNA methylation patterns, we generated 140 million additional RRBS reads (5.8 gigabase (Gb); Supplementary Information) from ES-derived neural precursor cells (NPCs) and various primary cell populations (Supplementary Table 2). We also generated new chromatin-state maps of H3 lysine 4 mono- and di-methylation (H3K4me1 and H3K4me2) from ES cells, NPCs and whole brain tissue (Supplementary Table 3 and Supplementary Information), using chromatin immunoprecipitation followed by high-throughput sequencing (ChIP-Seq)11.
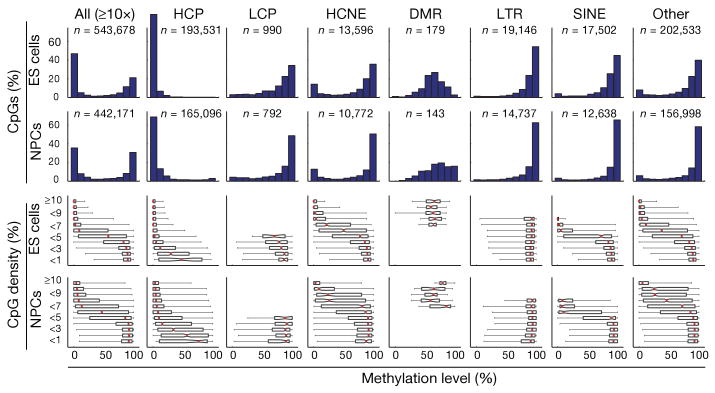
The methylation levels of CpG dinucleotides in wild-type ES cells display a bimodal distribution (Fig. 1), with most being either ‘largely unmethylated’ (<20% of reads showing methylation) or ‘largely methylated’ (>80% of reads). As expected2,8,12, CpGs in regions of high CpG density (>7% over 300 bp) tend to be unmethylated, whereas CpGs in low-density regions (<5%) tend to be methylated. However, we noted that ∼10% of CpGs in low-density regions were unmethylated, whereas ∼0.3% of CpGs in high-density regions were methylated. We found that DNA methylation patterns were better explained by histone methylation patterns than by CpG density. Because genomic features tend to be associated with distinct histone methylation patterns11, we analysed these features separately.
Figure 1. CpG methylation levels in ES cells and NPCs for CpGs with ≥10-fold coverage.
The top histograms show the distribution of methylation levels (%) across all CpGs, HCPs, LCPs, HCNEs, differentially methylated regions (DMRs), LTRs, SINEs and other genomic features (n, number of CpGs). Methylation levels are bimodal (except at DMRs, which have a unimodal distribution largely consistent with uniform sampling from the maternal and paternal alleles in ES cells and partial hypermethylation in NPCs). The bottom box plots show the distribution of methylation levels conditional on local CpG density (defined as fraction of CpGs in a 300-bp window; shown as percentage). The red lines denote medians, notches the standard errors, boxes the interquartile ranges, and whiskers the 2.5th and 97.5th percentiles.
High-CpG-density promoters (HCPs) are associated with two classes of genes: ubiquitous ‘housekeeping’ genes and highly regulated ‘key developmental’ genes13. In ES cells, HCPs at housekeeping genes are enriched with the transcription initiation mark H3K4me3 (‘univalent’) and are generally highly expressed, whereas those at developmental genes are enriched with both H3K4me3 and the repressive mark H3K27me3 (‘bivalent’) and are generally silent11,14. Both types of promoters are also enriched with H3K4me2, which is associated with an open chromatin confirmation. Out of the 10,299 HCPs sampled (on average, 19 distinct CpGs per promoter), we found that virtually all contain a core region of unmethylated CpGs, regardless of their level of expression or H3K27me3 enrichment (Figs 1 and 2a)12,14,15.
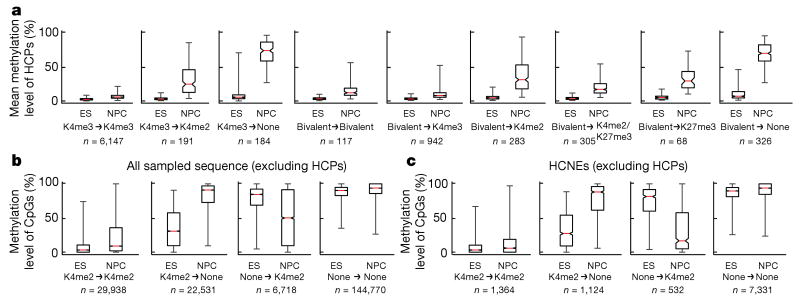
Figure 2. Correlation between DNA and histone methylation.
a, Mean methylation levels across CpGs within each profiled HCP (requiring ≥5-fold coverage of ≥5 CpGs), conditional on their histone methylation state in ES cells and NPCs (n, number of HCPs; those enriched with H3K4me3 are generally also enriched for H3K4me2, but not vice versa). Loss of H3K4 methylation, and to a lesser extent of H3K27me3, is correlated with gain of DNA methylation. b, Methylation levels of individual CpGs outside of HCPs, conditional on enrichment of H3K4me2 (n, number of distinct sites in each category). Changes in histone methylation state are inversely correlated with changes in DNA methylation. c, Methylation levels of CpGs in HCNEs not overlapping CpG islands, conditional on H3K4me2 enrichment. For a–c, the red lines denote medians, notches the standard errors, boxes the interquartile ranges, and whiskers the 2.5th and 97.5th percentiles. All pair-wise comparisons of methylation levels at sites with changing chromatin states are significant (P < 10−20, Mann–Whitney U test).
Low-CpG-density promoters (LCPs) are generally associated with tissue-specific genes. In ES cells, a small subset of LCPs are enriched with H3K4me3 (∼7%) or H3K4me2 (∼3%), and essentially none are enriched with H3K27me3 (ref. 11). We found that whereas most CpGs located in sampled LCPs (990 sites from 392 promoters) are methylated, those in LCPs enriched with H3K4me3 or H3K4me2 have significantly reduced methylation levels (Supplementary Fig. 4).
Distal regulatory regions such as enhancers, silencers and boundary elements are often required to establish correct gene expression patterns in mammalian cells16. Cis-regulatory elements active in a particular cell type are often associated with markers of open chromatin, such as H3K4me2 or H3K4me1 (refs 17, 18). We identified 25,051 sites of H3K4me2 enrichment in ES cells from 1 kb to >100 kb away from known promoters (most were also enriched with H3K4me1, but not with H3K4me3). CpGs sampled at H3K4me2-enriched sites (outside of promoters and CpG islands) had significantly lower methylation levels than those at unenriched sites (Fig. 2b). This relationship was particularly strong for CpGs located in highly conserved non-coding elements (HCNEs; Fig. 2c).
Imprinting control regions (ICRs) are CpG-rich regulatory regions that display allele-specific histone and DNA methylation19. Our RRBS library included sequences from 13 of ∼20 known ICRs (on average, 13 distinct CpGs per ICR). CpGs within these elements display a unimodal distribution of methylation levels, with a median close to 50%, which is consistent with hypomethylation of the active allele marked with H3K4me3 and hypermethylation of the silenced allele marked with H3K9me3 (Fig. 1)11.
Interspersed repeat families differ in their chromatin structure, with H3K9me3 enriched at active long terminal repeats (LTRs) and to a lesser extent at long interspersed elements (LINEs), but not at short interspersed elements (SINEs). Notably, CpGs located in LTRs and LINEs are generally hypermethylated, even in CpG-rich contexts (Fig. 1). In contrast, CpGs in SINEs show a correlation between methylation levels and CpG density that is comparable to non-repetitive sequences.
We conclude that in ES cells the presence of H3K4 methylation and the absence of H3K9 methylation are better predictors of unmethylated CpGs than sequence context alone. This is consistent with models in which de novo methyl-transferases either specifically recognize sites with unmethylated H3K4 (ref. 20) or are excluded by H3K4 methylation or associated factors. Similarly, H3K9me3 or associated factors may recruit methyl-transferases at ICRs and repetitive elements21.
We next used RRBS to analyse how DNA methylation patterns change when ES cells are differentiated in vitro into a homogeneous population of NPCs (Supplementary Fig. 4)22. Whereas CpG methylation levels are highly correlated between the two cell types (rho = 0.81), there were clear differences: ∼8% of CpGs unmethylated in ES cells became largely methylated in NPCs, whereas ∼2% of CpGs methylated in ES cells became unmethylated; these changes were strongly correlated with changes in histone methylation patterns.
At both univalent and bivalent HCPs, we found that most CpGs remained unmethylated on differentiation, particularly within their core CpG island, but that loss of H3K4me3 and retention of H3K4me2 or H3K27me3 correlated with a partial increase in DNA methylation levels (median, ∼25%; 2.9% and 32% of univalent and bivalent HCPs, respectively) and complete loss of H3K4 and H3K27 methylation correlated with DNA hypermethylation (median, ∼75%; 2.8% and 16% of univalent and bivalent HCPs, respectively; Fig. 2).
Most LCPs marked by H3K4 methylation in ES cells lose this mark in NPCs; however, LCPs associated with genes expressed in NPCs gain this mark. Loss or gain of H3K4 methylation is a strong predictor of inverse changes in CpG methylation levels at these promoters (Supplementary Fig. 5).
Our chromatin-state maps revealed that 18,899 (75%) of putative distal regulatory elements enriched with H3K4me2 in ES cells lost this mark in NPCs, whereas 20,088 new H3K4me2 sites appeared, often in HCNE-rich regions surrounding activated developmental genes (Fig. 3). Loss or gain of H3K4 methylation were again inversely correlated with CpG methylation levels (Fig. 2b, c). In fact, these regions account for most observed de-methylation events. The presence of H3K27me3 alone did not correlate with lower methylation levels in CpG-poor regions (Supplementary Fig. 6).
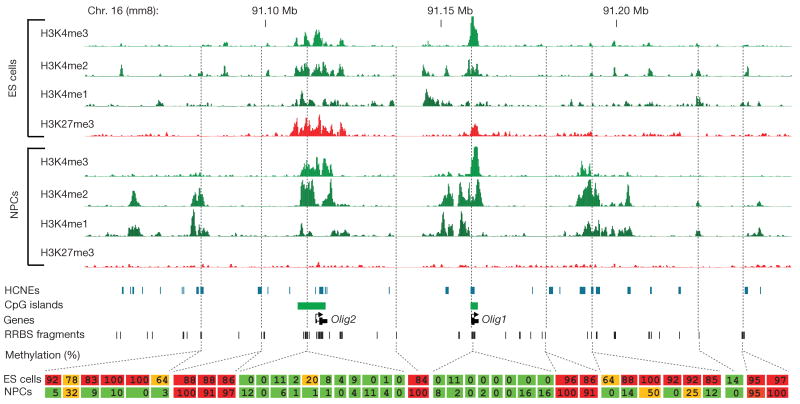
Figure 3. Developmentally regulated de-methylation of highly conserved non-coding elements.
Comparison of histone and DNA methylation levels across the Olig1/Olig2 neural-lineage transcription factor locus. ChIP-Seq tracks for H3K4me1/2/3 and H3K27me3 in ES cells and NPCs are shown. The unmethylated CpG-rich promoters are bivalent and inactive in ES cells and resolve to univalent H3K4me3 on activation in NPCs. H3K4me2 enrichment appears over HCNEs distal to the two genes, and this correlates with CpG de-methylation. Inferred methylation levels for 40 out of 215 sampled CpGs are shown and colour-coded. Red indicates largely methylated (>80%); green indicates largely unmethylated (<20%), and orange indicates intermediate levels (≥20% and ≤80%).
The data support the notion that CpG-rich and -poor regulatory elements undergo distinct modes of epigenetic regulation2,11,12. Most (>95%) HCPs seem to be constitutively unmethylated and regulated by trithorax-group (trxG; associated with H3K4me3) and/or Polycomb-group (PcG; associated with H3K27me3) proteins, which may be recruited in part by means of non-specific unmethylated-CpG binding domains23. Hypermethylation of these CpG-dense regions leads to exclusion of trxG/PcG activity, heterochromatin formation and essentially irreversible gene silencing2. In contrast, regulatory elements in CpG-poor sequence contexts seem to undergo extensive and dynamic methylation and de-methylation. Hence, methylation of isolated CpGs may contribute to chromatin condensation or directly interfere with transcription factor binding2, but does not necessarily prevent chromatin remodelling in response to activating signals.
As noted above, a small set of HCPs (n = 252; ∼3%) became hypermethylated (>75% mean methylation across sampled CpGs) on in vitro differentiation of ES cells to NPCs. To investigate whether the observed pattern reflects an in vivo regulatory mechanism, we isolated NPCs from embryonic day (E)13.5 embryos and differentiated them into glial fibrillary acidic protein (Gfap)-positive astrocytes (with no more than two passages in vitro). We similarly differentiated the in vitro-derived NPCs into astrocytes (with these cells having undergone at least 18 passages; Supplementary Fig. 4), and compared the two populations using RRBS (Fig. 4a–f).
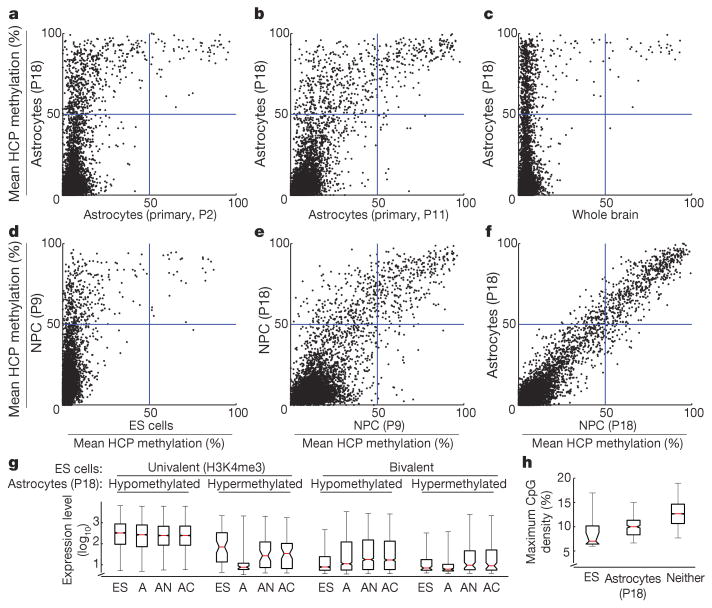
Figure 4. HCP hypermethylation of cultured cells.
Inferred mean methylation levels (%) across autosomal HCPs (requiring ≥5-fold coverage of ≥5 CpGs within the CpG island). a, ES-derived astrocytes contains roughly 10 times more hypermethylated HCPs than primary NPC-derived astrocytes after two passages (P) in culture. b, Continued passage of the primary cells lead to gradual hypermethylation of many of the same HCPs. c, Only a handful of mainly germline-specific HCPs display hypermethylation in a whole brain tissue sample. d, Most HCPs are unmethylated in ES cells, but a small subset gain significant methylation on differentiation to NPCs. e, Continued proliferation of NPCs leads to additional HCPs becoming hypermethylated after 18 passages. f, Differentiation of late-stage NPCs into astrocytes by growth factor withdrawal does not lead to additional HCP hypermethylation. g, Expression levels of genes associated with profiled HCPs for ES cells (ES), ES-derived astrocytes (A), primary neocortical astrocytes (AN) and cerebellar astrocytes (AC). Hypermethylation of HCPs is correlated with low expression levels in ES-derived astrocytes. HCPs that are univalent in ES cells and become hypermethylated in ES-derived astrocytes are associated with lower expression levels in both ES cells and primary astrocytes. h, The maximal CpG densities (300-bp window) of hypermethylated HCPs in ES cells or ES-derived astrocytes are significantly lower than for unmethylated HCPs. For g and h, the red lines denote medians, notches the standard errors, boxes the interquartile ranges, and whiskers the 2.5th and 97.5th percentiles.
The methylation levels of CpGs were highly correlated (rho = 0.85), but astrocytes obtained from in vivo NPCs displayed substantially less HCP hypermethylation than those obtained from ES cells (Fig. 4a). The in vivo-derived astrocytes showed hypermethylation at only 30 HCPs, largely associated with germline-specific genes (including Dazl, Hormad1, Sycp1, Sycp2 and Taf7l), several of which also showed partial methylation in ES cells. In contrast, the in vitro-derived astrocytes showed hypermethylation of these and ∼305 additional HCPs. This set includes some genes known to be expressed by at least some in vivo astrocytes (including Isyna1, Gsn and Cldn5; ref. 24) but that were silent in the ES-cell-derived astrocytes (Supplementary Information). However, the hypermethylated HCPs are significantly enriched for genes not expressed in NPCs or in the astrocyte lineage (Supplementary Tables 4–7). They include genes involved in development and differentiation of neuronal (Lhx8, Lhx9, Moxd1, Htr1f and Slit1), ependymal (Otx2 and Kl) and unrelated lineages (including Myod1, Dhh and Nkx3-1). In fact, we found that ‘key developmental’ HCPs that are bivalent in ES cells are six times more likely to be included in the hypermethylated set compared to univalent HCPs. Moreover, univalent genes in the hypermethylated set are expressed at significantly lower levels in both ES cells and primary astrocytes, compared to those that remained hypomethylated (Fig. 4g). We also found that the hypermethylated HCPs tend to have a ∼15% lower CpG density (Fig. 4h).
To investigate further the differences between in vitro and in vivo cell populations, we analysed whole brain tissue (representing cells of mainly glial origin). Virtually all (>99%) of sampled HCPs were unmethylated (Fig. 4c) and enriched with H3K4me3 and/or H3K27me3 (Supplementary Fig. 7), with ∼20 germline-specific HCPs being the only clear exceptions. RRBS libraries from other in vivo sources (T cells, B cells, spleen, lung, liver and fibroblasts) also showed few hypermethylated HCPs (Supplementary Fig. 8). This suggests that—apart from silencing germline-specific12, imprinted and X-inactivated (Supplementary Fig. 9) genes in somatic tissues—hypermethylation of HCPs is not a major mechanism of developmental regulation in vivo.
To test for a correlation between passage number and HCP hypermethylation, we examined independently derived in vitro NPCs collected after only 9 passages. These cells displayed hypermethylation at approximately half of the HCPs that are hypermethylated in the NPCs after 18 passages (Fig. 4d, e). To reduce time in culture further, we used Sox1–GFP (green fluorescent protein) ES cells25 to isolate very early NPCs. These cells initially displayed virtually no HCP hypermethylation. However, after continued culturing they acquired hypermethylation at many of the same HCPs as the previous NPC populations (Supplementary Fig. 8). Finally, we grew the in vivo-derived NPCs for 11 passages in vitro, differentiated them into astrocytes and then examined the methylation pattern. Notably, these cells had also begun to acquire hypermethylation at a largely similar set of HCPs (Fig. 4a, b).
These results show that independently derived NPC populations from both in vitro and in vivo sources and different genetic backgrounds reproducibly undergo gradual hypermethylation at a characteristic set of HCPs. These observations have several implications.
First, aberrant epigenetic regulation in culture has raised concern over the accuracy of cellular models generated by in vitro differentiation or manipulation26–28. Both primary and transformed cell lines, including ES-derived NPC populations, tend to lose developmental potency after continued proliferation in culture26,29. Susceptibility to hypermethylation at key regulatory genes that are normally activated on differentiation could explain this phenomenon. Second, malignant cells are often found to harbour hypermethylated CpG islands4,5. Recently, genes known to undergo frequent hypermethylation in adult cancers were noted to be significantly enriched for genes with bivalent promoters in ES cells (reviewed in ref. 30). The similarities between hypermethylation in culture and in cancer may provide a useful in vitro model for studying a common underlying mechanism. Finally, the gradual hypermethylation of ‘weak’ HCPs hints at underlying kinetics. Because H3K4 methylases are targeted, at least in part, by non-specific CpG-binding domains23, such HCPs may be particularly sensitive to imbalanced chromatin-modifying factors or other cancer- or culture-related perturbations.
More generally, RRBS makes it feasible to perform genome-scale bisulphite sequencing on large-mammalian genomes, providing a valuable tool for epigenetic profiling of cell populations. As sequencing capacity increases, genome coverage can be readily scaled in step by adding restriction enzymes, increasing the selected size range or using hybridization-based reduced representation strategies.
Methods Summary
ES cells and ES-derived neural cells were cultured as described previously11,25. Primary tissues were isolated from 4–6-week-old male 129SvJae/C57/B6 mice. Mouse embryonic fibroblasts (MEFs) and primary neural precursors were isolated from 129SvJae/C57/B6 E14.5 embryos.
RRBS libraries were prepared from 1–10 μg mouse genomic DNA digested with 10–100 Units MspI (NEB). Size-selected MspI fragments (40–120 bp and 120–220 bp) were filled in and 3′-terminal-A extended, extracted with phenol and precipitated with ethanol. Ligation to pre-annealed adapters containing 5′-methyl-cytosine instead of cytosine (Illumina) was performed using the Illumina DNA preparation kit and protocol. QIAquick (Qiagen) cleaned-up, adaptor-ligated fragments were bisulphite-treated using the EpiTect Bisulphite Kit (Qiagen). Preparative-scale PCR was performed and QIAquick-purified PCR products were subjected to a final size selection on a 4% NuSieve 3:1 agarose gel. SYBR-green-stained gel slices containing adaptor-ligated fragments of 130–210 bp or 210–310 bp in size were excised. Library material was recovered from the gel (QIAquick) and sequenced on an Illumina 1G genome analyser.
Sequence reads from bisulphite-treated Solexa libraries were identified using standard Illumina base-calling software and then analysed using a custom computational pipeline. ChIP-Seq experiments, sequencing, alignments and identification of significantly enriched regions were carried out as described previously11.
Supplementary Material
Acknowledgments
We thank the staff of the Broad Institute Genome Sequencing Platform for assistance with data generation and B. Ramsahoye for the nearest neighbour analysis. This research was supported by funds from the National Human Genome Research Institute, the National Cancer Institute, and the Broad Institute of MIT and Harvard.
Footnotes
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
Full Methods and any associated references are available in the online version of the paper at www.nature.com/nature.
References
- 1.Bestor TH. The DNA methyltransferases of mammals. Hum Mol Genet. 2000;9:2395–2402. doi: 10.1093/hmg/9.16.2395. [DOI] [PubMed] [Google Scholar]
- 2.Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002;16:6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- 3.Reik W. Stability and flexibility of epigenetic gene regulation in mammalian development. Nature. 2007;447:425–432. doi: 10.1038/nature05918. [DOI] [PubMed] [Google Scholar]
- 4.Feinberg AP. The epigenetics of cancer etiology. Semin Cancer Biol. 2004;14:427–432. doi: 10.1016/j.semcancer.2004.06.005. [DOI] [PubMed] [Google Scholar]
- 5.Jones PA, Baylin SB. The epigenomics of cancer. Cell. 2007;128:683–692. doi: 10.1016/j.cell.2007.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Meissner A, et al. Reduced representation bisulfite sequencing for comparative high-resolution DNA methylation analysis. Nucleic Acids Res. 2005;33:5868–5877. doi: 10.1093/nar/gki901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Frommer M, et al. A genomic sequencing protocol that yields a positive display of 5-methylcytosine residues in individual DNA strands. Proc Natl Acad Sci USA. 1992;89:1827–1831. doi: 10.1073/pnas.89.5.1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eckhardt F, et al. DNA methylation profiling of human chromosomes 6, 20 and 22. Nature Genet. 2006;38:1378–1385. doi: 10.1038/ng1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cokus SJ, et al. Shotgun bisulphite sequencing of the Arabidopsis genome reveals DNA methylation patterning. Nature. 2008;452:215–219. doi: 10.1038/nature06745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Altshuler D, et al. An SNP map of the human genome generated by reduced representation shotgun sequencing. Nature. 2000;407:513–516. doi: 10.1038/35035083. [DOI] [PubMed] [Google Scholar]
- 11.Mikkelsen TS, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Weber M, et al. Distribution, silencing potential and evolutionary impact of promoter DNA methylation in the human genome. Nature Genet. 2007;39:457–466. doi: 10.1038/ng1990. [DOI] [PubMed] [Google Scholar]
- 13.Saxonov S, Berg P, Brutlag DL. A genome-wide analysis of CpG dinucleotides in the human genome distinguishes two distinct classes of promoters. Proc Natl Acad Sci USA. 2006;103:1412–1417. doi: 10.1073/pnas.0510310103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bernstein B, et al. A bivalent chromatin structure marks key Developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- 15.Illingworth R, et al. A novel CpG island set identifies tissue-specific methylation at developmental gene loci. PLoS Biol. 2008;6:e22. doi: 10.1371/journal.pbio.0060022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.West AG, Fraser P. Remote control of gene transcription. Hum Mol Genet. 2005;14(Spec No 1):R101–111. doi: 10.1093/hmg/ddi104. [DOI] [PubMed] [Google Scholar]
- 17.Heintzman ND, et al. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nature Genet. 2007;39:311–318. doi: 10.1038/ng1966. [DOI] [PubMed] [Google Scholar]
- 18.Bernstein BE, et al. Genomic maps and comparative analysis of histone modifications in human and mouse. Cell. 2005;120:169–181. doi: 10.1016/j.cell.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 19.Edwards CA, Ferguson-Smith AC. Mechanisms regulating imprinted genes in clusters. Curr Opin Cell Biol. 2007;19:281–289. doi: 10.1016/j.ceb.2007.04.013. [DOI] [PubMed] [Google Scholar]
- 20.Ooi SK, et al. DNMT3L connects unmethylated lysine 4 of histone H3 to de novo methylation of DNA. Nature. 2007;448:714–717. doi: 10.1038/nature05987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Esteve PO, et al. Direct interaction between DNMT1 and G9a coordinates DNA and histone methylation during replication. Genes Dev. 2006;20:3089–3103. doi: 10.1101/gad.1463706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Conti L, et al. Niche-independent symmetrical self-renewal of a mammalian tissue stem cell. PLoS Biol. 2005;3:e283. doi: 10.1371/journal.pbio.0030283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Voo KS, Carlone DL, Jacobsen BM, Flodin A, Skalnik DG. Cloning of a mammalian transcriptional activator that binds unmethylated CpG motifs and shares a CXXC domain with DNA methyltransferase, human trithorax, and methyl-CpG binding domain protein 1. Mol Cell Biol. 2000;20:2108–2121. doi: 10.1128/mcb.20.6.2108-2121.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sharma MK, et al. Distinct genetic signatures among pilocytic astrocytomas relate to their brain region origin. Cancer Res. 2007;67:890–900. doi: 10.1158/0008-5472.CAN-06-0973. [DOI] [PubMed] [Google Scholar]
- 25.Aubert J, et al. Screening for mammalian neural genes via fluorescence-activated cell sorter purification of neural precursors from Sox1–gfp knock-in mice. Proc Natl Acad Sci USA. 2003;100(Suppl 1):11836–11841. doi: 10.1073/pnas.1734197100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jones PA, Wolkowicz MJ, Harrington MA, Gonzales F. Methylation and expression of the Myo D1 determination gene. Phil Trans R Soc Lond B. 1990;326:277–284. doi: 10.1098/rstb.1990.0011. [DOI] [PubMed] [Google Scholar]
- 27.Smiraglia DJ, et al. Excessive CpG island hypermethylation in cancer cell lines versus primary human malignancies. Hum Mol Genet. 2001;10:1413–1419. doi: 10.1093/hmg/10.13.1413. [DOI] [PubMed] [Google Scholar]
- 28.Shen Y, Chow J, Wang Z, Fan G. Abnormal CpG island methylation occurs during in vitro differentiation of human embryonic stem cells. Hum Mol Genet. 2006;15:2623–2635. doi: 10.1093/hmg/ddl188. [DOI] [PubMed] [Google Scholar]
- 29.Bouhon IA, Joannides A, Kato H, Chandran S, Allen ND. Embryonic stem cell-derived neural progenitors display temporal restriction to neural patterning. Stem Cells. 2006;24:1908–1913. doi: 10.1634/stemcells.2006-0031. [DOI] [PubMed] [Google Scholar]
- 30.Ohm JE, Baylin SB. Stem cell chromatin patterns: an instructive mechanism for DNA hypermethylation? Cell Cycle. 2007;6:1040–1043. doi: 10.4161/cc.6.9.4210. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.