Abstract
Stable isotope labeling by amino acids in cell culture (SILAC) provides a straightforward tool for quantitation in proteomics. However, one problem associated with SILAC is the in vivo conversion of labeled arginine to other amino acids, typically proline. We found that arginine conversion in the fission yeast Schizosaccharomyces pombe occurred at extremely high levels, such that labeling cells with heavy arginine led to undesired incorporation of label into essentially all of the proline pool as well as a substantial portion of glutamate, glutamine, and lysine pools. We found that this can be prevented by deleting genes involved in arginine catabolism using methods that are highly robust yet simple to implement. Deletion of both fission yeast arginase genes or of the single ornithine transaminase gene, together with a small modification to growth medium that improves arginine uptake in mutant strains, was sufficient to abolish essentially all arginine conversion. We demonstrated the usefulness of our approach in a large scale quantitative analysis of proteins before and after cell division; both up- and down-regulated proteins, including a novel protein involved in septation, were successfully identified. This strategy for addressing the “arginine conversion problem” may be more broadly applicable to organisms amenable to genetic manipulation.
Stable isotope labeling by amino acids in cell culture (SILAC)1 (1) is one of the key methods for large scale quantitative proteomics (2, 3). In SILAC experiments, proteins are metabolically labeled by culturing cells in media containing either normal (“light”) or heavy isotope-labeled amino acids, typically lysine and arginine. Peptides derived from the light and heavy cells are thus distinguishable by mass spectrometry and can be mixed for accurate quantitation. SILAC is also possible at the whole-organism level (4).
An inherent problem in SILAC is the metabolic conversion of labeled arginine to other amino acids, as this complicates quantitative analysis of peptides containing these amino acids. Arginine conversion to proline is well described in mammalian cells, although the extent of conversion varies among cell types (5). When conversion is observed, typically 10–25% of the total proline pool is found to contain label (6–11). Arginine conversion has also been reported in SILAC experiments with budding yeast Saccharomyces cerevisiae (3, 12, 13).
Because more than 50% of tryptic peptides in large data sets contain proline (7), it is not practical simply to disregard proline-containing peptides during quantitation. Several methods have been proposed to either reduce arginine conversion or correct for its effects on quantitation. In some cell types, arginine conversion can be prevented by lowering the concentration of exogenous arginine (6, 14–16) or by adding exogenous proline (9). However, these methods can involve significant changes to growth media and may need to be tested for each experimental condition used. Given the importance of arginine in many metabolic pathways, careful empirical titration of exogenous arginine concentration is required to minimize negative effects on cell growth (14). In addition, low arginine medium can lead to incomplete arginine labeling, although the reasons for this are not entirely clear (7). An alternative strategy is to omit labeled arginine altogether (3, 13, 17), but this reduces the number of quantifiable peptides. Correction methods include using two different forms of labeled arginine (7) or computationally compensating for proline-containing peptides (11, 12, 18). Ultimately, none of these methods address the problem at its root, the utilization of arginine in cellular metabolism.
To develop a differential proteomics work flow for the fission yeast Schizosaccharomyces pombe, we sought to adapt SILAC for use in this organism, a widely used model eukaryote with excellent classical and reverse genetics. Here we describe extremely high conversion of labeled arginine to other amino acids in fission yeast as well as a novel general solution to the problem that should be applicable to other organisms. As proof of principle, we quantitated changes in protein levels before and after cell division on a proteome-wide scale. We identified both up- and down-regulated proteins, including a novel protein involved in septation.
EXPERIMENTAL PROCEDURES
Yeast Methods
S. pombe methods were performed as described, and Edinburgh minimal medium 2 (EMM2) using phthalate buffer as specified by Nurse and co-workers (19) was used throughout. car1Δ, aru1Δ, and car2Δ single deletion strains (for ORFs SPBP26C9.02c, SPAC3H1.07, and SPBC21C3.08c, respectively) were purchased from Bioneer and crossed into laboratory strains as required. A green fluorescent protein (GFP)-tagged Uds1 (SPBC27.04) strain was constructed by PCR-based gene targeting as described (20). A list of yeast strains used is shown in supplemental Table 1. For MS analysis of amino acid conversion, cells were grown to mid-log phase in EMM2 with the relevant amino acid supplements, then diluted into EMM2 containing either l-[13C6]arginine·HCl (=“[13C6]arginine”) or l[13C6]lysine·2HCl (=“[13C6]lysine”; both from Cambridge Isotope Laboratories) at either 20 or 120 mg/liter, and grown at 30 °C for seven generations to a final A595 of 0.8 (107 cells/ml) before harvesting (see below).
Because labeled amino acids are expensive, we determined optimal concentrations for their use in fission yeast, in relation to cost, by measuring terminal ODs of cultures of arg1–230 lys3–37 car2Δ auxotrophs grown in different combinations of arginine and lysine concentrations. When arginine concentration was 40 mg/liter, increases in lysine concentration beyond 30 mg/liter did not increase the terminal OD. Similarly, when lysine concentration was 30 mg/liter, increases in arginine concentration beyond 40 mg/liter did not increase the terminal OD. We therefore normally supplemented EMM2 with arginine at 40 mg/liter and lysine at 30 mg/liter so that neither amino acid was in excess. Doubling the concentrations to 80 mg/liter arginine and 60 mg/liter lysine (which maintains the 4:3 weight ratio of arginine:lysine) approximately doubled the terminal OD, but further increases in concentrations gave less proportionate increases in OD. We thus recommend using arginine and lysine at this 4:3 weight ratio in the concentration range described.
For G2 arrest-and-release experiments, cdc25–22 nmt81:GFP-atb2 arg1–230 lys3–37 car2Δ cells were grown in EMM2 using 6 mm ammonium chloride as nitrogen source and supplemented with 40 mg/liter [12C6]arginine and [12C6]lysine or [13C6]arginine and [13C6]lysine. (The cdc25–22 arg1–230 lys3–37 car2Δ mutant showed increased sensitivity to ammonium chloride as compared with arg1–230 lys3–37 car2Δ cells, which grew well in ammonium chloride concentrations up to 24 mm. When in doubt, lower ammonium chloride concentrations are preferred.) Cells were grown at 25 °C for seven generations until A595 0.2 (2.5 × 106 cells/ml) at which time 11 mm hydroxyurea was added to the cultures. After 5 h, cells were washed with prewarmed medium at 36 °C and shifted to 36 °C for 3 h 40 min. Cells were then shifted back to 25 °C, and samples were collected every 10 min for 180 min. Mitotic stages (i.e. prometaphase, metaphase, and anaphase) were assayed by fluorescence microscopy of GFP-tubulin (21), and septation index was monitored by staining with Tinopal UNPA-GX (Sigma F-3543) and fluorescence microscopy. Percentages of cells in different stages at different times were converted into normalized cumulative data for presentation in Fig. 6B.
Fig. 6.
G2 arrest-and-release experiment using cdc25–22 mutant. A, experimental design. cdc25–22 nmt81:GFP-atb2 arg1–230 lys3–37 car2Δ cells are partially synchronized in G1/S by hydroxyurea (HU) treatment and washout, shifted up to non-permissive temperature to allow accumulation in late G2, and then shifted down to permissive temperature for a synchronous mitosis. Experimental manipulations are shown in green, and cell states are shown in black. The midpoint in septation (10hr10′) corresponds approximately to G1/S because G1 is very short in fission yeast, and by the time daughter cells are physically separated (septation complete), cells are in G2 (DNA synthesis is essentially complete). Asynchr., asynchronous. B, percentages of cells that have entered prometaphase/metaphase, anaphase (assayed by GFP-tubulin fluorescence), or septation (assayed by Tinopal UNPA-GX staining) after shifting down from G2 arrest.
Protein Methods
Protein extraction methods were identical for assaying amino acid incorporation and for G2 arrest-and-release experiments. Cells were harvested for 4 min at 4000 rpm at 20 °C, washed once with TBS, and centrifuged for 1 min at 13,000 rpm. The cell pellet was resuspended in a small volume of TBS, incubated at 95 °C for 5 min to inactivate proteases, and disrupted by bead beating with 0.5 mm zirconia beads in a Ribolyser (Hybaid) at room temperature using two cycles of 30 s at the maximum setting (“6.5”). Disrupted cells were extracted by addition of an equal volume of 2× Laemmli sample buffer (0.125 m Tris, pH 6.8, 4% SDS, 20% glycerol without dye or reducing agent) and incubation at 95 °C for 5 min. The extract was then centrifuged for 10 min at 13,000 rpm at room temperature. The supernatant was recovered, and protein concentration was determined using the bicinchoninic acid assay. Extracts were supplemented with bromphenol blue and reducing agent prior to electrophoresis.
For analysis of amino acid conversion, 100 μg of protein extract was electrophoresed by SDS-PAGE, and a region of the gel containing a major Coomassie-staining band was excised and then subjected to further manipulation as described below. For analysis of G2 arrest and release, 300 μg of protein extract was briefly electrophoresed by SDS-PAGE and excised without fractionation for further manipulation as described below.
For Western blotting of Cdc13 and Uds1-GFP protein levels, 50 μg of total cell extract (per lane) was electrophoresed by 8% SDS-PAGE and transferred to nitrocellulose in 10 mm 3-(cyclohexylamino)-1-propanesulfonic acid, pH 11 with 10% methanol. The membrane was blocked with TBS containing 2% nonfat milk and 0.2% Tween 20. Primary antibody incubations were done overnight at 4 °C in the same buffer. Monoclonal anti-Cdc13 clone 6F11/2 (Cancer Research UK) was used at a concentration of 10 μg/ml. Rabbit anti-Bip1 was a kind gift from Alison Pidoux (University of Edinburgh) and was used at 1:10,000 dilution of serum. Sheep affinity-purified anti-GFP was generated in house and used at 20 μg/ml. Membranes were washed 3 × 10 min in TBS containing 0.02% Tween 20 and incubated with secondary antibody for 2 h at room temperature. Secondary antibodies were IRDye 800 donkey anti-mouse, IRDye 680 donkey anti-rabbit, and IRDye 800 donkey anti-goat (LI-COR Biosciences). Membranes were washed 3 × 5 min with TBS containing 0.02% Tween 20, and a final wash was done with TBS only. Proteins were detected with an Odyssey scanner (LI-COR Biosciences), and intensity of the bands was quantified with Odyssey software (version 3.0). Cdc13 and Uds1-GFP signals were normalized against Bip1, which does not vary in levels during the cell cycle.2
Sample Preparation and Fractionation for Mass Spectrometry
For analysis of arginine conversion, a band of Coomassie-stained gel (200 μg of total protein extract loaded) was excised, and proteins were digested using trypsin at an enzyme-to-protein ratio of 1:50 as described (22). In brief, proteins were reduced in 10 mm DTT for 30 min at 37 °C, alkylated in 55 mm iodoacetamide for 20 min at room temperature in the dark, and digested overnight at 37 °C with 12.5 ng/μl trypsin (proteomics grade, Sigma). The digestion medium was then acidified, and peptides were desalted on StageTips as described (23, 24) for LC-MS/MS analysis.
For G2 arrest-and-release experiments, the gel piece containing the entire protein sample was treated as described for arginine conversion. Four separate MS analyses were performed involving three independent biological replicates. Different (i.e. reversed) labeling and fractionation strategies were used as shown in supplemental Table 2.
For strong cation exchange fractionation, sample digestion was performed as described above and quenched with phosphoric acid (Sigma-Aldrich) until pH 3.0 was reached. After recovery of peptides, the gel piece was re-extracted with 100% acetonitrile, and the two recoveries were pooled. The sample was then diluted with Buffer A (5 mm KH2PO4, 10% acetonitrile, pH 3.0) to 20% acetonitrile and fractionated using strong cation exchange chromatography (2.1-mm × 20-cm polysulfoethyl A column, Poly LC, Columbia, MD) on an UltiMate 3000 HPLC instrument (Dionex UK Ltd., Surrey, UK). Peptides were separated using Buffer A, Buffer B (Buffer A with 1 m KCl), a flow rate of 200 μl/min, and a 50-min gradient as follows: 0–1 min, 0% Buffer B; 1–5 min, raise to 0.5% Buffer B; 5–14 min, raise to 1.0% Buffer B; 14–41 min, raise to 70% Buffer B (curve 7 equation, CHROMELEON software version 6.80, Dionex UK Ltd.); 41–42 min, 70% Buffer B. Fractions were collected every 1 min. All the fractions were diluted with 0.1% TFA at a ratio of 1:1 and desalted using StageTips for subsequent LC-MS/MS analysis.
For OFFGEL fractionation, the digestion was performed as described above, acidified with 0.1% TFA, and desalted using StageTips. The peptide samples were mixed with peptide OFFGEL stock solution (glycerol + OFFGEL Buffer, high resolution kit, Agilent). IPG strip pH 3–10 24-cm strip/24-well frame was used. The fractionation was performed following manufacturer's instructions (OG24PE00, Agilent 3100 OFFGEL fractionator). Once the fractionation was finished, the status of the machine changed from focusing to hold. At this point, the 24 fractions were collected and desalted using StageTips.
Mass Spectrometric Analysis
An LTQ-Orbitrap mass spectrometer (ThermoElectron) was coupled on line to an Agilent 1100 binary nanopump and an HTC PAL autosampler (CTC Analytics). To prepare an analytical column with a self-assembled particle frit (25), C18 material (ReproSil-Pur C18-AQ 3 μm, Dr. Maisch, GmbH) was packed into a spray emitter (75-μm inner diameter, 8-μm opening, 70-mm length; New Objectives) using an air pressure pump (Proxeon Biosystems). Mobile phase A consisted of water, 5% acetonitrile, and 0.5% acetic acid. Mobile phase B consisted of acetonitrile and 0.5% acetic acid. The gradient used for each fraction varied from 2 to 4 h depending on the estimated concentration of peptides in the fraction. Peptides were loaded onto the column at a flow rate of 0.7 μl/min and eluted at a flow rate of 0.3 μl/min. For a 2-h gradient run, elution used a gradient from 0% B to 20% B over 75 min and then from 20% B to 80% B over 13 min. Fourier transform mass spectrometry spectra were recorded at 30,000 resolution, and the six most intense peaks of the MS scan were selected in the ion trap for fragmentation (normal scan; wideband activation; filling, 7.5 × 105 ions for MS scan and 1.5 × 104 ions for MS/MS; maximum fill time, 150 ms; dynamic exclusion for 60 s). Raw files were processed using MaxQuant software (version 1.0.13.8) (26) at default parameters except that labels “lysine-6” and “arginine-6” were selected.
Searches were conducted using Mascot (version 2.2.0) against a concatenated target-decoy database containing S. pombe sequences (S. pombe GeneDB, downloaded December 17, 2009, 5003 sequences) to which we added the sequences of porcine trypsin, trypsinogen anionic precursor, and bovine serum albumin. The decoy part of the database was obtained by inverting the target sequences. The search parameters were as follows: mass tolerance for precursor ions, 6 ppm; mass tolerance for fragment ions, 0.5 Da; enzyme specificity, fully tryptic (no cleavage N-terminally of proline); allowed number of missed cleavages, 2; fixed modification, carbamidomethylation on cysteine; variable modification, oxidation on methionine and N-acetylation on protein N terminus. 13C6 label on lysine and arginine was set as fixed or variable modification or not used depending on which MaxQuant peak list was searched (26). Identifications and SILAC ratios of proteins were obtained using MaxQuant at default parameters, including identification confidence 99% at peptide and protein levels and requiring a minimum of two quantified SILAC pairs for quantitation. For protein identification, two peptides were required, among which at least one peptide was required to be unique in the database. If a group of identified peptide sequences belonged to multiple proteins and these proteins could not be distinguished, i.e. no unique peptide was reported, these proteins were reported as a protein group by MaxQuant.
To generate a list of quantified proteins for the final data set shown in Fig. 7A, data from the four separate MS analyses were combined in MaxQuant, but to qualify for entry into this list, a given protein had to be independently quantified by MaxQuant (i.e. not merely identified) in individual analyses of at least two of the three independent biological replicates described above. This filter reduced the total number of proteins from 3575 (total combined identified, including not quantified; supplemental Table 4) to 2964 (total combined quantified; supplemental Table 5).
Fig. 7.
Changes in fission yeast protein levels during progression from late G2 to G1/S. A, log2 protein expression ratios (G1/S versus late G2) for 2964 fission yeast proteins (x axis) plotted against the sum of the relevant peptide intensities (y axis). Proteins are colored according to values of MaxQuant Significance(B) (26); gray, Significance(B) > 10−2; orange, 10−3 < Significance(B) < 10−2; blue, 10−5 < Significance(B) < 10−3; red, Significance(B) < 10−5. Proteins with Significance(B) < 10−3 that are previously described as being cell cycle-regulated are indicated in black. A previously uncharacterized protein, Uds1 (SPBC27.04), is indicated in green. B, Western blots of GFP-tagged Uds1 and B-type cyclin Cdc13 after release from G2 arrest. Bip1 is a loading control. C, quantitation of Uds1-GFP and Cdc13 levels from B.
Gene Ontology Classification
Biological process gene ontology (GO) terms were assigned to each up-regulated protein, and the highest level GO term in the ontology (i.e. a broad description of protein function) was chosen for each protein. The assigned GO terms were grouped to correspond to general biological categories (shown in supplemental Table 3). The number of total-quantified proteins with the chosen GO terms was obtained from the Gene Ontology web site. This was then used to determine the proportional representation of the relevant biological categories in the up-regulated proteins versus the total quantified proteins.
Live Cell Imaging
Uds1-GFP cells were grown in EMM2 at 25 °C and imaged at room temperature. Images were collected on a Nikon TE2000 inverted microscope coupled to a Yokogawa CSU-10 spinning disc confocal head (Visitech) and an IxonEM+ DU-888 electron-multiplied charge-coupled device camera (Andor). Samples were illuminated with a 488 nm Coherent 20-milliwatt laser operating at 15–20% intensity. Eight Z-sections at 0.6-μm spacing were acquired every 2 min for 46 min and then every 3 min for the next 69 min. Image acquisition, processing, and analysis were carried out using Metamorph software (Molecular Devices). Maximum projections are shown.
RESULTS
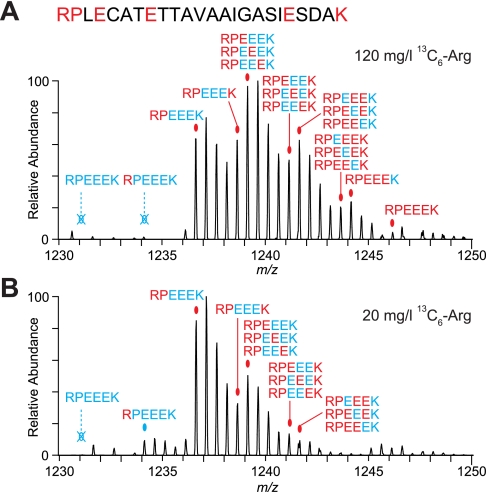
Growth of fission yeast lys3–37 lysine auxotrophs in 13C6-labeled lysine led to incorporation of label on lysine and not on other amino acids (supplemental Fig. S1). However, when arg1–230 arginine auxotrophs were grown in 13C6-labeled arginine, complex isotope clusters were observed, indicating extensive arginine conversion (Fig. 1A). Significant conversion occurred even with levels of exogenous arginine that are too low to support robust growth (Fig. 1B; additional data not shown). Further analysis revealed incorporation of label in all detectable proline as well as in 25–30% of glutamate, glutamine, and lysine (Figs. 1 and 2); no other amino acids were labeled (Fig. 2 and supplemental Fig. S2). To our knowledge, this is the most extreme example of arginine conversion observed in SILAC labeling experiments to date, and it severely compromises proteomics analyses. In particular, simultaneous partial incorporation of label into multiple amino acids not only makes it highly challenging to implement computational correction methods but also can compromise peptide identification itself as a result of reduced success in identifying monoisotopic peaks and increased complexity of fragmentation spectra (Fig. 3, C and D). Furthermore, large and complicated isotope clusters challenge dynamic exclusion during data acquisition, which subsequently reduces the number of different peptides selected for fragmentation.
Fig. 1.
Extensive conversion of labeled arginine to other amino acids in fission yeast. A and B, mass spectra of peptide RPLECATETTAVAAIGASIESDAK from S. pombe pyruvate kinase (SPAC4H3.10c) isolated from arg1–230 cells grown in 120 mg/liter 13C6-labeled arginine (A) or 20 mg/liter 13C6-labeled arginine (B). At the lower arginine concentration, arg1–230 mutants grow only to a low density. Red letters in the peptide sequence show amino acids observed to incorporate label. Red ovals indicate monoisotopic peaks of differently labeled versions of the peptide containing labeled amino acids other than arginine. Hollow blue crossed out ovals indicate the absence of peaks containing either no label or only labeled arginine. The blue oval indicates a monoisotopic peak of the version of the peptide in which only arginine is labeled. In peak annotations, the peptide sequence was reduced to the potentially labeled amino acids; red denotes labeled amino acids, and blue denotes unlabeled amino acids.
Fig. 2.
Arginine is converted to proline, glutamate, glutamine, and lysine. Shown are mass spectra of peptides isolated from S. pombe arg1–230 cells grown in 120 mg/liter 13C6-labeled arginine. A and B, peptide containing a single proline and its fragmentation spectrum, showing full labeling of proline. C–E, peptides containing a single glutamine (C), glutamate (D), or lysine (E) showing 25–30% labeling of these amino acids. Figure annotation is as in Fig. 1.
Fig. 3.
Multiple partially labeled amino acids create complex peptide spectra. A–C, peptides from S. pombe arg1–230 cells grown in 120 mg/liter 13C6-labeled arginine, showing increasing complexity of spectra when multiple amino acids are partially labeled. D, fragmentation spectrum of m/z 942.04 for the peptide shown in C (shaded), revealing complex peak patterns in which differently labeled versions of equivalent fragments are spaced by 1, 4, or 5 Da depending on the combination of labeled amino acids present in the respective versions (expanded region, below). Note that selecting m/z 942.04 will include arginine-labeled ALIMDLEGLTGDIVKR that contains additional label not only on either glutamine or glutamate but also alternatively on lysine (m/z 941.54) due to the width of the selection window for fragmentation. Figure annotation is as in Fig. 1.
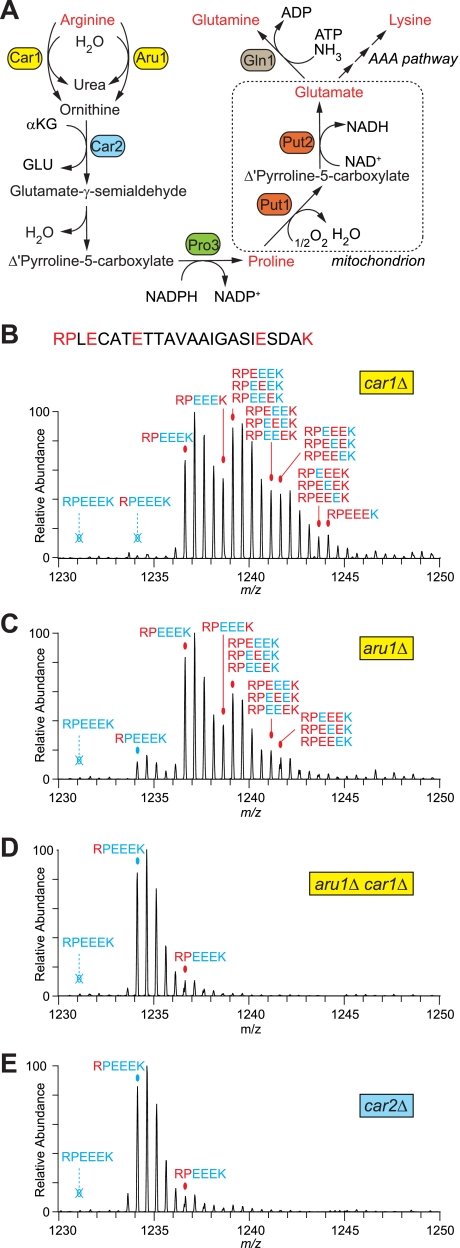
We reasoned that such extreme levels of arginine conversion might be most effectively prevented by deleting genes involved in arginine catabolism. Fig. 4A shows expected pathways in fission yeast leading from arginine to proline and glutamate, and ultimately to glutamine and lysine, based on work in budding yeast and filamentous fungi (27–29). We focused our attention on the arginases Car1 (30) and Aru1 and on ornithine transaminase Car2. In car1Δ and aru1Δ single deletion strains, labeled arginine was still converted to other amino acids, although conversion was less extreme in aru1Δ mutants than in car1Δ mutants (Fig. 4, B and C). This suggests that Aru1 is the major arginase in fission yeast. Interestingly, however, conversion was almost completely prevented in a car1Δ aru1Δ double deletion strain (Fig. 4D). In a car2Δ single deletion strain, arginine conversion was similarly prevented (Fig. 4E). We used the car2Δ strain in subsequent work because only a single mutation was needed to prevent conversion.
Fig. 4.
Genetic manipulation of arginine catabolism prevents arginine conversion. A, metabolic pathways leading to conversion of arginine to other amino acids in fungi. “AAA pathway” indicates the α-aminoadipate pathway, which is fed by equilibration of glutamate (GLU) with α-ketoglutarate (αKG) (29). B–E, mass spectra of the same peptide, RPLECATETTAVAAIGASIESDAK, shown in Fig. 1 isolated from arg1–230 car1Δ (B), arg1–230 aru1Δ (C), arg1–230 aru1Δ car1Δ (D), and arg1–230 car2Δ (E) cells, all grown in 120 mg/liter 13C6-labeled arginine. Figure annotation is as in Fig. 1.
Complete incorporation of labeled amino acids in SILAC experiments requires growth for at least six or seven cell generations under labeling conditions. To our surprise, when lysine and arginine auxotrophies were combined in a single strain, dilution of exponentially growing cells for labeling purposes led to an extremely long lag phase, lasting several days, prior to resumption of exponential growth (Fig. 5). By contrast, wild-type prototrophs resumed exponential growth immediately. Previous work has suggested the existence of at least two distinct systems for arginine uptake in fission yeast, one that is generic for basic amino acids and thus can be competitively inhibited by lysine and a second that is arginine-specific but is inhibited by ammonium chloride, the nitrogen source in the standard fission yeast minimal growth medium EMM2 (19, 31). The combination of lys3–37 and arg1–230 auxotrophies in a single strain thus poses a particularly difficult challenge for arginine uptake and robust growth. To address this problem, we asked whether lowering the ammonium chloride concentration of EMM2 might allow the arg1–230 lys3–37 car2Δ strain to return more rapidly to exponential growth. Strikingly, we found that a 4–16-fold reduction of ammonium chloride led to an immediate return of this strain to exponential growth with generation times and terminal cell densities nearly as high as those in wild-type cells (Fig. 5). These results indicate that a slightly modified version of EMM2, with no observable adverse effects on cell growth, is highly suitable for SILAC in fission yeast.
Fig. 5.
Lowering ammonium chloride concentration allows rapid return of arg1–230 lys3–37 mutants to exponential growth after dilution. Shown are growth curves of wild-type and arg1–230 lys3–37 car2Δ cells grown to exponential phase in EMM2 (plus exogenous arginine and lysine) at the different ammonium chloride concentrations shown and then diluted in the same medium at time 0.
To validate these approaches in a proof-of-principle SILAC experiment, we used our mutant strains and modified growth medium to measure protein levels before and after cell division on a proteome-wide scale. Cells from a cdc25–22 G2 arrest-and-release experiment (32) were harvested at the time of release (late G2 phase) and at the peak of septation index (corresponding to G1/S phase), by which time essentially all cells had progressed through the metaphase-anaphase transition (Fig. 6). From an analysis of three independent biological replicates by LC-MS/MS and MaxQuant software (26), we quantitated levels in late G2 versus G1/S for 2964 proteins of an estimated maximal 5027 possible fission yeast ORFs (Fig. 7A and supplemental Tables 4–6). Because only 80–90% of fission yeast genes are thought to be expressed in vegetative cells (33, 34), our quantitative analysis likely covered ∼70% of the expressed proteome. To our knowledge, this is the first large scale SILAC analysis in fission yeast following earlier label-free approaches (35) and illustrates the usefulness of the method.
We found that 2805 proteins (∼95% of total quantified) did not change significantly in levels (Fig. 7A and supplemental Fig. S3). Sixty-five proteins were down-regulated between late G2 and G1/S with high statistical confidence; most of these showed a relatively small -fold change in levels (Fig. 7A and supplemental Table 5). The majority of down-regulated proteins (∼70%) have not been experimentally characterized. Based on gene ontology annotation, the most common biological function associated with the down-regulated proteins is “cellular stress response” with ∼30% of the characterized and uncharacterized proteins associated with this grouping (data not shown). The greatest -fold down-regulated protein was the B-type cyclin protein Cdc13, which is degraded at the metaphase-anaphase transition by the anaphase-promoting complex (APC) (32, 36–38). We confirmed the extent of Cdc13 down-regulation by quantitative Western blotting (Fig. 7, B and C).
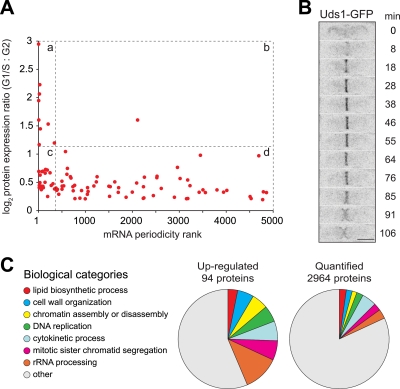
We identified 94 proteins up-regulated between late G2 and G1/S (supplemental Table 5); five of these are already known to vary at the protein level (Fig. 7A) (39–43). We further validated our results by comparing them with previous transcriptome analyses (44–48). Approximately 35 of the 94 up-regulated proteins were up-regulated transcriptionally at this stage of the cell cycle, and of the 10 most up-regulated proteins in our experiments, nine showed very strong transcriptional periodicity and phasing consistent with our results (Fig. 8A, quadrant a). This suggests that large increases in protein expression between G2 and G1/S in fission yeast are mostly due to transcriptional rather than post-transcriptional mechanisms (Fig. 8A, compare quadrants a and b). Another ∼20 proteins had a significantly lower degree of up-regulation in SILAC but still showed periodic transcription of the corresponding genes (Fig. 8A, quadrant c). For many of these, the peaks and troughs of transcription may be out of phase with our experimental sampling (47) so that actual -fold changes in protein expression may be higher than those we measured.
Fig. 8.
Highly up-regulated proteins exhibit strong periodicity of mRNA expression. A, log2 protein expression ratios (G1/S versus G2) for the 94 identified up-regulated proteins plotted against the periodicity rank of the corresponding mRNAs (data from Cyclebase (48)). Dashed lines indicate quadrants described in text. Although periodicity is a continuous variable, mRNAs to the left of the dashed vertical line are widely acknowledged as periodic; beyond rank 500, mRNAs are generally considered non-periodic (47). B, images from time lapse fluorescence videomicroscopy showing the appearance of Uds1-GFP at the cell division site during septation in an asynchronous culture. Minutes of total elapsed time are shown at right. Bar, 5 μm. C, relative proportion of the 94 up-regulated proteins represented in selected biological categories compared with representation of the 2964 quantified proteins.
Compared with the 2964 quantified proteins, the 94 up-regulated proteins were enriched for gene ontology terms relating to cell division and cell wall/septum organization (Fig. 8C). The highest -fold up-regulated protein was the uncharacterized ORF SPBC27.04, which we named Uds1 (“up-regulated during septation”). We followed expression and localization of Uds1 through the cell cycle by tagging the corresponding gene with GFP at its chromosomal locus. In G2 arrest-and-release experiments, we measured a 5-fold increase in expression of Uds1-GFP between late G2 and G1/S by quantitative Western blotting (Fig. 7, B and C). In actively cycling cells, we observed that Uds1-GFP appears during the onset of septation, localizes uniquely to the septum, and then disappears with ensuing cell separation (Fig. 8B). Uds1 is thus a novel fission yeast protein involved in septation.
DISCUSSION
The fission yeast S. pombe is an important model organism for many aspects of eukaryotic cell biology, in particular cell cycle regulation and cell division (49). Several aspects of fission yeast cell biology that are shared with multicellular eukaryotes are not present in the budding yeast S. cerevisiae, including distributed centromeres, RNA interference, HP1-dependent heterochromatin, and certain aspects of telomere regulation, as well as cell cycle-dependent changes in microtubule dynamics and microtubule regulation of cell polarity. Given the powerful genetic tools available for fission yeast, it is of considerable importance to develop a robust SILAC platform for this organism. Large scale data sets for fission yeast proteomics have been generated only relatively recently (for example, see Refs. 35, 50, and 51), and proteomics analyses using SILAC in fission yeast have until now not been reported.
We found that labeled arginine in fission yeast was converted not only extensively to proline but also significantly to glutamate, glutamine, and lysine, posing a major challenge for SILAC experiments in fission yeast. By contrast, in mammalian cells, arginine conversion to amino acids other than proline has not been observed, although it is theoretically possible (1, 9, 11). The very high degree of conversion to proline in fission yeast is likely responsible for this difference. In addition, in mammalian cells one would not expect conversion from arginine to lysine, as mammals lack the α-aminoadipate pathway found in higher fungi and must obtain lysine from diet (or culture medium) (29). The genetic engineering solution to the arginine conversion problem presented in this work may be particularly appropriate in cases where conversion is extremely high, as lowering the exogenous arginine concentration was not sufficient to prevent conversion in our experiments, although it is often sufficient in mammalian cells (see the Introduction).
Although nearly all arginine conversion was prevented in aru1Δ car1Δ and car2Δ strains, a very low amount of conversion was nevertheless present, to proline only (Fig. 4, D and E). We estimate this to represent less than 10% of the proline pool, and as it would have only negligible effects in our cell cycle experiments, this low level of conversion was not taken into account. If necessary, a further degree of accuracy in future fission yeast SILAC experiments might be achieved by a small computational correction (11, 12, 18).
In contrast to lowering the arginine concentration, which can be deleterious to cell growth and/or physiology (Refs. 7 and 52) and our data), our approach to preventing arginine conversion in fission yeast enables the use of labeled arginine without any suboptimal modifications to the growth medium, in this case EMM2, the defined standard minimal medium for physiological experiments in fission yeast (19). Although we lowered the concentration of ammonium chloride in conventional EMM2 to improve arginine uptake in arg1–230 lys3–37 mutants, this had no adverse effect on cell growth or generation times. Indeed, even in low ammonium medium, supplemented arginine and lysine were still limiting for growth (data not shown), indicating that the ammonium chloride in the original EMM2 formulation is likely present in considerable excess to what is actually necessary.
Applying our fission yeast SILAC methodology in a proof-of-principle experiment, we were able to identify both proteins that were up-regulated and proteins that were down-regulated as cells progressed from G2 to G1/S, and we showed that the most up-regulated protein in our data set, Uds1, is a novel protein that transiently localizes to the septum during cell division. Among the down-regulated proteins, only the B-type cyclin Cdc13 has been previously described to be cell cycle-regulated at the protein level (32, 36–38). Like Cdc13, the fission yeast securin protein Cut2 is degraded at the metaphase-anaphase transition via the APC (53). However, Cut2 was not present in our data set, and thus, we cannot judge whether we would have detected it as a down-regulated protein. Slp1 (Cdc20 homolog), an activator of the APC, is also degraded, later in anaphase, by the APC (54). Slp1 was also not present in our data set, but it is tightly regulated at both the protein and transcript levels and increases only transiently during preanaphase mitosis. Thus, even if detected, Slp1 would not have appeared as down-regulated between G2 and G1/S. This highlights the importance of temporal sampling frequency and phasing in experiments that seek to comprehensively identify cell cycle-regulated changes in protein levels. In budding yeast, the microtubule-bundling protein Ase1p is described as being degraded in anaphase by APC (55). We identified fission yeast Ase1 (SPAPB1A10.09) in our analysis, but it was not significantly changed in levels between G2 and G1/S in our experiments (ratio G1/S:G2 = 0.96; supplemental Table 5). This may reflect the different biology of the two yeasts, as fission yeast Ase1 is important for microtubule bundling in interphase as well as in mitosis (56, 57), and thus its disappearance at the end of mitosis could be detrimental to the cell. A more complete description of down-regulated proteins will require a fully comprehensive identification of the fission yeast proteome involving more in-depth analysis and/or additional methods such as selected reaction monitoring techniques (58). Indeed, in our view, our results do not provide a basis for estimating how many down-regulated proteins might be found in such an analysis as protein identifications based on MS tend to discriminate against very low abundance proteins. Low abundance proteins are likely to be significantly under-represented in our data set of quantified proteins, which contains 2964 of the estimated 4000–4500 proteins thought to be expressed in vegetative cells (33, 34).
In summary, although our approach was motivated by the very high levels of arginine conversion seen in fission yeast relative to mammalian cells, it is likely that as SILAC becomes more widely adopted in other organisms similarly high levels of arginine conversion will be encountered elsewhere. Where cells are amenable to genetic manipulation, targeting of arginase and/or ornithine transaminase genes should provide a useful solution. Gene deletion strains in many microorganisms are relatively easy to generate and widely used. For example, in budding yeast, there are single genes for both arginase (CAR1) and ornithine transaminase (CAR2), and deletions in both produce viable cells (59). In addition, in organisms where genetic manipulation is possible but less straightforward, one could nevertheless take advantage of a genetic engineering solution to arginine conversion by constructing a reference strain that can be heavy isotope-labeled and then used as an internal standard in relation to non-engineered light isotope-labeled strains, similar to SILAC work with tissue samples (60, 61).
Supplementary Material
Acknowledgments
This work was a collaboration of the Sawin and Rappsilber laboratories. We thank J. Zou for help with sample fractionation, J.-C. Bukowksi-Wills and A. Kerr for help with data analysis, A. Pidoux for anti-Bip1 antibody, and P. Fantes for discussions about amino acid uptake.
* This work was supported in part by the Wellcome Trust, the Fundação para a Ciência e Tecnologia (FCT), and the European Commission.
 This article contains supplemental Tables 1–6 and Figs. S1–S3.
This article contains supplemental Tables 1–6 and Figs. S1–S3.
2 A. Pidoux, personal communication.
1 The abbreviations used are:
- SILAC
- stable isotope labeling by amino acids in cell culture
- EMM2
- Edinburgh minimal medium 2
- GFP
- green fluorescent protein
- GO
- gene ontology
- APC
- anaphase-promoting complex.
REFERENCES
- 1.Ong S. E., Blagoev B., Kratchmarova I., Kristensen D. B., Steen H., Pandey A., Mann M. (2002) Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol. Cell. Proteomics 1, 376–386 [DOI] [PubMed] [Google Scholar]
- 2.Ong S. E., Mann M. (2005) Mass spectrometry-based proteomics turns quantitative. Nat. Chem. Biol. 1, 252–262 [DOI] [PubMed] [Google Scholar]
- 3.de Godoy L. M., Olsen J. V., Cox J., Nielsen M. L., Hubner N. C., Fröhlich F., Walther T. C., Mann M. (2008) Comprehensive mass-spectrometry-based proteome quantification of haploid versus diploid yeast. Nature 455, 1251–1254 [DOI] [PubMed] [Google Scholar]
- 4.Krüger M., Moser M., Ussar S., Thievessen I., Luber C. A., Forner F., Schmidt S., Zanivan S., Fässler R., Mann M. (2008) SILAC mouse for quantitative proteomics uncovers kindlin-3 as an essential factor for red blood cell function. Cell 134, 353–364 [DOI] [PubMed] [Google Scholar]
- 5.Ong S. E., Kratchmarova I., Mann M. (2003) Properties of 13C-substituted arginine in stable isotope labeling by amino acids in cell culture (SILAC). J. Proteome Res. 2, 173–181 [DOI] [PubMed] [Google Scholar]
- 6.Blagoev B., Mann M. (2006) Quantitative proteomics to study mitogen-activated protein kinases. Methods 40, 243–250 [DOI] [PubMed] [Google Scholar]
- 7.Van Hoof D., Pinkse M. W., Oostwaard D. W., Mummery C. L., Heck A. J., Krijgsveld J. (2007) An experimental correction for arginine-to-proline conversion artifacts in SILAC-based quantitative proteomics. Nat. Methods 4, 677–678 [DOI] [PubMed] [Google Scholar]
- 8.Schmidt F., Strozynski M., Salus S. S., Nilsen H., Thiede B. (2007) Rapid determination of amino acid incorporation by stable isotope labeling with amino acids in cell culture (SILAC). Rapid Commun. Mass Spectrom. 21, 3919–3926 [DOI] [PubMed] [Google Scholar]
- 9.Bendall S. C., Hughes C., Stewart M. H., Doble B., Bhatia M., Lajoie G. A. (2008) Prevention of amino acid conversion in SILAC experiments with embryonic stem cells. Mol. Cell. Proteomics 7, 1587–1597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liao L., Park S. K., Xu T., Vanderklish P., Yates J. R., 3rd (2008) Quantitative proteomic analysis of primary neurons reveals diverse changes in synaptic protein content in fmr1 knockout mice. Proc. Natl. Acad. Sci. U.S.A. 105, 15281–15286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Park S. K., Liao L., Kim J. Y., Yates J. R., 3rd (2009) A computational approach to correct arginine-to-proline conversion in quantitative proteomics. Nat. Methods 6, 184–185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gruhler A., Olsen J. V., Mohammed S., Mortensen P., Faergeman N. J., Mann M., Jensen O. N. (2005) Quantitative phosphoproteomics applied to the yeast pheromone signaling pathway. Mol. Cell. Proteomics 4, 310–327 [DOI] [PubMed] [Google Scholar]
- 13.Soufi B., Kelstrup C. D., Stoehr G., Fröhlich F., Walther T. C., Olsen J. V. (2009) Global analysis of the yeast osmotic stress response by quantitative proteomics. Mol. Biosyst. 5, 1337–1346 [DOI] [PubMed] [Google Scholar]
- 14.Ong S. E., Mann M. (2006) A practical recipe for stable isotope labeling by amino acids in cell culture (SILAC). Nat. Protoc. 1, 2650–2660 [DOI] [PubMed] [Google Scholar]
- 15.Graumann J., Hubner N. C., Kim J. B., Ko K., Moser M., Kumar C., Cox J., Schöler H., Mann M. (2008) Stable isotope labeling by amino acids in cell culture (SILAC) and proteome quantitation of mouse embryonic stem cells to a depth of 5,111 proteins. Mol. Cell. Proteomics 7, 672–683 [DOI] [PubMed] [Google Scholar]
- 16.Prokhorova T. A., Rigbolt K. T., Johansen P. T., Henningsen J., Kratchmarova I., Kassem M., Blagoev B. (2009) Stable isotope labeling by amino acids in cell culture (SILAC) and quantitative comparison of the membrane proteomes of self-renewing and differentiating human embryonic stem cells. Mol. Cell. Proteomics 8, 959–970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hwang S. I., Lundgren D. H., Mayya V., Rezaul K., Cowan A. E., Eng J. K., Han D. K. (2006) Systematic characterization of nuclear proteome during apoptosis: a quantitative proteomic study by differential extraction and stable isotope labeling. Mol. Cell. Proteomics 5, 1131–1145 [DOI] [PubMed] [Google Scholar]
- 18.Mousson F., Kolkman A., Pijnappel W. W., Timmers H. T., Heck A. J. (2008) Quantitative proteomics reveals regulation of dynamic components within TATA-binding protein (TBP) transcription complexes. Mol. Cell. Proteomics 7, 845–852 [DOI] [PubMed] [Google Scholar]
- 19.Moreno S., Klar A., Nurse P. (1991) Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 194, 795–823 [DOI] [PubMed] [Google Scholar]
- 20.Bähler J., Wu J. Q., Longtine M. S., Shah N. G., McKenzie A., 3rd, Steever A. B., Wach A., Philippsen P., Pringle J. R. (1998) Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast 14, 943–951 [DOI] [PubMed] [Google Scholar]
- 21.Sawin K. E., Lourenco P. C., Snaith H. A. (2004) Microtubule nucleation at non-spindle pole body microtubule-organizing centers requires fission yeast centrosomin-related protein mod20p. Curr. Biol. 14, 763–775 [DOI] [PubMed] [Google Scholar]
- 22.Shevchenko A., Wilm M., Vorm O., Mann M. (1996) Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal. Chem. 68, 850–858 [DOI] [PubMed] [Google Scholar]
- 23.Rappsilber J., Ishihama Y., Mann M. (2003) Stop and go extraction tips for matrix-assisted laser desorption/ionization, nanoelectrospray, and LC/MS sample pretreatment in proteomics. Anal. Chem. 75, 663–670 [DOI] [PubMed] [Google Scholar]
- 24.Rappsilber J., Mann M., Ishihama Y. (2007) Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using StageTips. Nat. Protoc. 2, 1896–1906 [DOI] [PubMed] [Google Scholar]
- 25.Ishihama Y., Rappsilber J., Andersen J. S., Mann M. (2002) Microcolumns with self-assembled particle frits for proteomics. J. Chromatogr. A 979, 233–239 [DOI] [PubMed] [Google Scholar]
- 26.Cox J., Mann M. (2008) MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26, 1367–1372 [DOI] [PubMed] [Google Scholar]
- 27.Jones E. W., Fink G. R. (1982) The regulation of amino acid and nucleotide biosynthesis in yeast, in The Molecular Biology of the Yeast Saccharomyces: Vol. II. Metabolism and Gene Expression (Strathern J. N., Jones E. W., Broach J. R. eds) pp. 181–299, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 28.Davis R. H. (1986) Compartmental and regulatory mechanisms in the arginine pathways of Neurospora crassa and Saccharomyces cerevisiae. Microbiol. Rev. 50, 280–313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xu H., Andi B., Qian J., West A. H., Cook P. F. (2006) The alpha-aminoadipate pathway for lysine biosynthesis in fungi. Cell Biochem. Biophys. 46, 43–64 [DOI] [PubMed] [Google Scholar]
- 30.Van Huffel C., Dubois E., Messenguy F. (1994) Cloning and sequencing of Schizosaccharomyces pombe car1 gene encoding arginase. Expression of the arginine anabolic and catabolic genes in response to arginine and related metabolites. Yeast 10, 923–933 [DOI] [PubMed] [Google Scholar]
- 31.Fantes P. A., Creanor J. (1984) Canavanine resistance and the mechanism of arginine uptake in the fission yeast Schizosaccharomyces pombe. J. Gen. Microbiol. 130, 3265–3273 [DOI] [PubMed] [Google Scholar]
- 32.Moreno S., Hayles J., Nurse P. (1989) Regulation of p34cdc2 protein kinase during mitosis. Cell 58, 361–372 [DOI] [PubMed] [Google Scholar]
- 33.Lyne R., Burns G., Mata J., Penkett C. J., Rustici G., Chen D., Langford C., Vetrie D., Bähler J. (2003) Whole-genome microarrays of fission yeast: characteristics, accuracy, reproducibility, and processing of array data. BMC Genomics 4, 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wilhelm B. T., Marguerat S., Watt S., Schubert F., Wood V., Goodhead I., Penkett C. J., Rogers J., Bähler J. (2008) Dynamic repertoire of a eukaryotic transcriptome surveyed at single-nucleotide resolution. Nature 453, 1239–1243 [DOI] [PubMed] [Google Scholar]
- 35.Schmidt M. W., Houseman A., Ivanov A. R., Wolf D. A. (2007) Comparative proteomic and transcriptomic profiling of the fission yeast Schizosaccharomyces pombe. Mol. Syst. Biol. 3, 79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Booher R. N., Alfa C. E., Hyams J. S., Beach D. H. (1989) The fission yeast cdc2/cdc13/suc1 protein kinase: regulation of catalytic activity and nuclear localization. Cell 58, 485–497 [DOI] [PubMed] [Google Scholar]
- 37.Yamano H., Gannon J., Hunt T. (1996) The role of proteolysis in cell cycle progression in Schizosaccharomyces pombe. EMBO J. 15, 5268–5279 [PMC free article] [PubMed] [Google Scholar]
- 38.Tanaka K., Petersen J., MacIver F., Mulvihill D. P., Glover D. M., Hagan I. M. (2001) The role of Plo1 kinase in mitotic commitment and septation in Schizosaccharomyces pombe. EMBO J. 20, 1259–1270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Birkenbihl R. P., Subramani S. (1995) The rad21 gene product of Schizosaccharomyces pombe is a nuclear, cell cycle-regulated phosphoprotein. J. Biol. Chem. 270, 7703–7711 [DOI] [PubMed] [Google Scholar]
- 40.Buck V., Ng S. S., Ruiz-Garcia A. B., Papadopoulou K., Bhatti S., Samuel J. M., Anderson M., Millar J. B., McInerny C. J. (2004) Fkh2p and Sep1p regulate mitotic gene transcription in fission yeast. J. Cell Sci. 117, 5623–5632 [DOI] [PubMed] [Google Scholar]
- 41.Martín-Cuadrado A. B., Dueñas E., Sipiczki M., Vázquez de Aldana C. R., del Rey F. (2003) The endo-beta-1,3-glucanase eng1p is required for dissolution of the primary septum during cell separation in Schizosaccharomyces pombe. J. Cell Sci. 116, 1689–1698 [DOI] [PubMed] [Google Scholar]
- 42.Pinar M., Coll P. M., Rincón S. A., Pérez P. (2008) Schizosaccharomyces pombe Pxl1 is a paxillin homologue that modulates Rho1 activity and participates in cytokinesis. Mol. Biol. Cell 19, 1727–1738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tasto J. J., Morrell J. L., Gould K. L. (2003) An anillin homologue, Mid2p, acts during fission yeast cytokinesis to organize the septin ring and promote cell separation. J. Cell Biol. 160, 1093–1103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rustici G., Mata J., Kivinen K., Lió P., Penkett C. J., Burns G., Hayles J., Brazma A., Nurse P., Bähler J. (2004) Periodic gene expression program of the fission yeast cell cycle. Nat. Genet. 36, 809–817 [DOI] [PubMed] [Google Scholar]
- 45.Oliva A., Rosebrock A., Ferrezuelo F., Pyne S., Chen H., Skiena S., Futcher B., Leatherwood J. (2005) The cell cycle-regulated genes of Schizosaccharomyces pombe. PLoS Biol. 3, e225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Peng X., Karuturi R. K., Miller L. D., Lin K., Jia Y., Kondu P., Wang L., Wong L. S., Liu E. T., Balasubramanian M. K., Liu J. (2005) Identification of cell cycle-regulated genes in fission yeast. Mol. Biol. Cell 16, 1026–1042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Marguerat S., Jensen T. S., de Lichtenberg U., Wilhelm B. T., Jensen L. J., Bähler J. (2006) The more the merrier: comparative analysis of microarray studies on cell cycle-regulated genes in fission yeast. Yeast 23, 261–277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gauthier N. P., Jensen L. J., Wernersson R., Brunak S., Jensen T. S. (2010) Cyclebase.org: version 2.0, an updated comprehensive, multi-species repository of cell cycle experiments and derived analysis results. Nucleic Acids Res. 38, D699–D702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nurse P. M. (2002) Nobel Lecture. Cyclin dependent kinases and cell cycle control. Biosci. Rep. 22, 487–499 [DOI] [PubMed] [Google Scholar]
- 50.Wilson-Grady J. T., Villén J., Gygi S. P. (2008) Phosphoproteome analysis of fission yeast. J. Proteome Res. 7, 1088–1097 [DOI] [PubMed] [Google Scholar]
- 51.Brill L. M., Motamedchaboki K., Wu S., Wolf D. A. (2009) Comprehensive proteomic analysis of Schizosaccharomyces pombe by two-dimensional HPLC-tandem mass spectrometry. Methods 48, 311–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wheatley D. N., Scott L., Lamb J., Smith S. (2000) Single amino acid (arginine) restriction: growth and death of cultured HeLa and human diploid fibroblasts. Cell Physiol. Biochem. 10, 37–55 [DOI] [PubMed] [Google Scholar]
- 53.Funabiki H., Yamano H., Kumada K., Nagao K., Hunt T., Yanagida M. (1996) Cut2 proteolysis required for sister-chromatid separation in fission yeast. Nature 381, 438–441 [DOI] [PubMed] [Google Scholar]
- 54.Yamada H. Y., Matsumoto S., Matsumoto T. (2000) High dosage expression of a zinc finger protein, Grt1, suppresses a mutant of fission yeast slp1(+), a homolog of CDC20/p55CDC/Fizzy. J. Cell Sci. 113, 3989–3999 [DOI] [PubMed] [Google Scholar]
- 55.Juang Y. L., Huang J., Peters J. M., McLaughlin M. E., Tai C. Y., Pellman D. (1997) APC-mediated proteolysis of Ase1 and the morphogenesis of the mitotic spindle. Science 275, 1311–1314 [DOI] [PubMed] [Google Scholar]
- 56.Yamashita A., Sato M., Fujita A., Yamamoto M., Toda T. (2005) The roles of fission yeast ase1 in mitotic cell division, meiotic nuclear oscillation, and cytokinesis checkpoint signaling. Mol. Biol. Cell 16, 1378–1395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Loïodice I., Staub J., Setty T. G., Nguyen N. P., Paoletti A., Tran P. T. (2005) Ase1p organizes antiparallel microtubule arrays during interphase and mitosis in fission yeast. Mol. Biol. Cell 16, 1756–1768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Picotti P., Bodenmiller B., Mueller L. N., Domon B., Aebersold R. (2009) Full dynamic range proteome analysis of S. cerevisiae by targeted proteomics. Cell 138, 795–806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Giaever G., Chu A. M., Ni L., Connelly C., Riles L., Véronneau S., Dow S., Lucau-Danila A., Anderson K., André B., Arkin A. P., Astromoff A., El-Bakkoury M., Bangham R., Benito R., Brachat S., Campanaro S., Curtiss M., Davis K., Deutschbauer A., Entian K. D., Flaherty P., Foury F., Garfinkel D. J., Gerstein M., Gotte D., Güldener U., Hegemann J. H., Hempel S., Herman Z., Jaramillo D. F., Kelly D. E.., Kelly S. L., Kötter P., LaBonte D., Lamb D. C., Lan N., Liang H., Liao H., Liu L., Luo C., Lussier M., Mao R., Menard P., Ooi S. L., Revuelta J. L., Roberts C. J., Rose M., Ross-Macdonald P., Scherens B., Schimmack G., Shafer B., Shoemaker D. D., Sookhai-Mahadeo S., Storms R. K., Strathern J. N., Valle G., Voet M., Volckaert G., Wang C. Y., Ward T. R., Wilhelmy J., Winzeler E. A., Yang Y., Yen G., Youngman E., Yu K., Bussey H., Boeke J. D., Snyder M., Philippsen P., Davis R. W., Johnston M. (2002) Functional profiling of the Saccharomyces cerevisiae genome. Nature 418, 387–391 [DOI] [PubMed] [Google Scholar]
- 60.Ishihama Y., Sato T., Tabata T., Miyamoto N., Sagane K., Nagasu T., Oda Y. (2005) Quantitative mouse brain proteomics using culture-derived isotope tags as internal standards. Nat. Biotechnol. 23, 617–621 [DOI] [PubMed] [Google Scholar]
- 61.Geiger T., Cox J., Ostasiewicz P., Wisniewski J. R., Mann M. (2010) Super-SILAC mix for quantitative proteomics of human tumor tissue. Nat. Methods 7, 383–385 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.