Preface
Antibiotic drug-target interactions, and their respective direct effects, are generally well-characterized. In contrast, the bacterial responses to antibiotic drug treatments that contribute to cell death are not as well understood and have proven to be quite complex, involving multiple genetic and biochemical pathways. Here, we review the multi-layered effects of drug-target interactions, including the essential cellular processes inhibited by bactericidal antibiotics and the associated cellular response mechanisms that contribute to killing by bactericidal antibiotics. We also discuss new insights into these mechanisms that have been revealed through the study of biological networks, and describe how these insights, together with related developments in synthetic biology, may be exploited to create novel antibacterial therapies.
Introduction
Our understanding of how antibiotics induce bacterial cell death is centered on the essential cellular function inhibited by the primary drug-target interaction. Antibiotics can be classified based on the cellular component or system they affect, in addition to whether they induce cell death (bactericidal drugs) or merely inhibit cell growth (bacteriostatic drugs). Most current bactericidal antimicrobials, which are the focus of this review, inhibit DNA synthesis, RNA synthesis, cell wall synthesis, or protein synthesis1.
Since the discovery of penicillin was reported in 19292, other, more effective antimicrobials have been discovered and developed by elucidation of drug-target interactions, and by drug molecule modification. These efforts have significantly enhanced our clinical armamentarium. Antibiotic-mediated cell death, however, is a complex process that begins with the physical interaction between a drug molecule and its bacterial-specific target, and involves alterations to the affected bacterium at the biochemical, molecular and ultrastructural levels. The increasing prevalence of drug-resistant bacteria3, as well as the means of gaining resistance, has made it crucial that we better understand the multilayered mechanisms by which currently available antibiotics kill bacteria, as well as explore and find alternative antibacterial therapies.
Antibiotic-induced cell death has been associated with the formation of double-stranded DNA breaks following treatment with DNA gyrase inhibitors4, with the arrest of DNA-dependent RNA synthesis following treatment with rifamycins5, with cell envelope damage and loss of structural integrity following treatment with cell-wall synthesis inhibitors6, and with cellular energetics, ribosome binding and protein mistranslation following treatment with protein synthesis inhibitors7. Additionally, recent evidence points toward a common mechanism of cell death, involving disadvantageous cellular responses to drug-induced stresses that are shared by all classes of bactericidal antibiotics, which ultimately contributes to killing by these drugs8. More specifically, treatment with lethal concentrations of bactericidal antibiotics results in the production of harmful hydroxyl radicals through a common oxidative damage cellular death pathway involving alterations in central metabolism (TCA cycle) and iron metabolism8–10.
Here we review our current knowledge of the drug-target interactions and associated mechanisms by which the major classes of bactericidal antibiotics kill bacteria. We also describe recent efforts in network biology that have yielded novel, mechanistic insights into how bacteria respond to lethal antibiotic treatments, and discuss how these insights and related developments in synthetic biology may be used to develop new, effective means to combat bacterial infections.
Inhibition of DNA replication by quinolones
Modulation of chromosomal supercoiling through topoisomerase-catalyzed strand breakage and rejoining reactions is required for DNA synthesis, mRNA transcription and cell division11–13. These reactions are exploited by the synthetic quinolone class of antimicrobials, including the clinically-relevant fluoroquinolones, which target DNA-topoisomerase complexes4, 14, 15. Quinolones are derivatives of nalidixic acid, which was discovered as a byproduct of chloroquine (quinine) synthesis and introduced in the 1960s to treat urinary tract infections16. Nalidixic acid and other first generation quinolones (i.e., oxolinic acid) are rarely used today owing to their toxicity17. Second (i.e., ciprofloxacin), third (i.e., levofloxacin) and fourth (i.e., gemifloxacin) generation quinolone antibiotics (Table 1) can be classified based on their chemical structure along with qualitative differences in how these drugs kill bacteria16, 18.
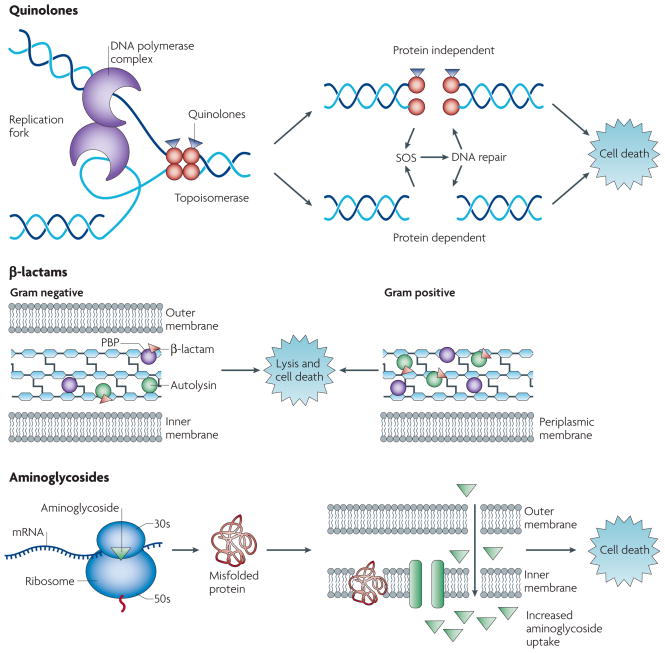
The quinolone class of antimicrobials interferes with the maintenance of chromosomal topology by targeting DNA gyrase (topoisomerase II) and topoisomerase IV (topoIV), trapping these enzymes at the DNA cleavage stage and preventing strand rejoining4, 19, 20 (Figure 1a). Despite the general functional similarities between topoIV and gyrase, the susceptibility of these targets to quinolone antibiotics varies across bacterial species20 (Table 1). For example, several studies have shown that topoIV is the primary target of quinolones in Gram-positive bacteria (e.g., Streptococcus pneumoniae21), whereas gyrase is the primary target and topoIV the secondary target of these drugs in Gram-negative bacteria (e.g., E. coli13 and Neisseria gonorrhoea22).
Figure 1. Drug-target interactions and associated cell death mechanisms.
a) Quinolone antibiotics interfere with changes in DNA supercoiling by binding to topoisomerase II or IV. This leads to the formation of double-stranded DNA breaks and cell death in either a protein synthesis dependent or protein synthesis independent fashion. b) β-lactams inhibit transpeptidation by binding to PBPs on maturing peptidoglycan strands. The decrease in peptidoglycan synthesis and increase in autolysins leads to lysis and cell death. c) Aminoglycosides bind to the 30S subunit of the ribosome and cause misincorporation of amino acids into elongating peptides. These mistranslated proteins can misfold, and incorporation of misfolded membrane proteins into the cell envelope leads to increased drug uptake, which together with an increase in ribosome binding has been associated with cell death.
Introduction of DNA breaks and replication fork arrest
The ability of quinolone antibiotics to kill bacteria is a function of the stable interaction complex formed between drug-bound topoisomerase enzyme and cleaved DNA4. Mechanistically, based on studies employing DNA cleavage mutants of gyrase23 and topoIV24 that do not prevent quinolone binding, as well as studies that have shown that strand breakage can occur in the presence of quinolones25, it is accepted that DNA strand breakage occurs after the drug has bound the enzyme. Therefore, the net effect of quinolone treatment is to generate double-stranded DNA breaks that are trapped by covalently (yet reversibly) linked topoisomerases whose functions are compromised26–28. As a result of quinolone-topoisomerase-DNA complex formation, DNA replication machinery becomes arrested at blocked replication forks, leading to inhibition of DNA synthesis, which immediately leads to bacteriostasis and eventually cell death4 (Figure 1a). It should be noted, however, that these effects on DNA replication can be correlated with bacteriostatic concentrations of quinolones, and are regarded as being reversible4, 29. Nonetheless, considering that gyrase has been found to be distributed approximately every 100 kilobases along the chromosome30, poisoning of topoisomerases by quinolone antibiotics and the resulting formation of stable complexes with DNA have substantial, negative consequences for the cell in terms of its ability to deal with drug-induced DNA damage31.
The role of protein expression in quinolone-mediated cell death
The introduction of double-stranded DNA breaks following topoisomerase inhibition by quinolones induces the DNA stress response (SOS response), in which RecA is activated by DNA damage and promotes auto-cleavage of the LexA repressor protein, inducing expression of SOS-response genes including DNA repair enzymes32. Notably, several studies have shown that preventing induction of the SOS response serves to enhance killing by quinolone antibiotics (except in the case of the first generation quinolone, nalidixic acid)8, 33. Preventing induction of the SOS response has also been shown to reduce the formation of drug-resistant mutants by blocking the induction of error-prone DNA polymerases34, homologous recombination20, and horizontal transfer of drug-resistance elements35, 36.
Together with studies revealing that co-treatment with quinolones and the protein synthesis inhibitor, chloramphenicol, inhibits the ability of certain quinolones to kill bacteria19, 37, there seems to be a clear relationship between the primary effects of quinolone-topoisomerase-DNA complex formation and the response of the bacteria (through the stress-induced expression of proteins) to these effects in the bactericidal activity of quinolone antibiotics. For example, ROS-mediated cell death has recently been shown to occur through the protein synthesis dependent pathway38. Also, the chromsomally-encoded toxin, MazF, has been shown to be required under certain conditions for efficient killing by quinolone drugs owing to its ability to alter protein carbonylation39, a form of oxidative stress40.
Inhibition of RNA synthesis by rifamycins
The inhibition of RNA synthesis by the rifamycin class of semi-synthetic bactericidal antibiotics, much like the inhibition of DNA replication by quinolones, has a catastrophic effect on prokaryotic nucleic acid metabolism and is a potent means for inducing bacterial cell death5. Rifamycin drugs inhibit DNA-dependent transcription by stable binding, with high affinity, to the subunit (encoded by the rpoB gene) of a DNA-bound and actively-transcribing RNA polymerase enzyme 41–43 (Table 1); the subunit is located within the channel formed by the polymerase-DNA complex, from which the newly synthesized RNA strand emerges 44. A unique mechanistic requirement of rifamycins is that RNA synthesis has not progressed beyond the addition of two ribonucleotides, which is attributed to the ability of the drug molecule to sterically inhibit nascent RNA strand initialization 45. It is worth noting that rifamycins are not thought to act by blocking the elongation step of RNA synthesis, although a recently discovered class of RNA polymerase inhibitors (based on the compound CBR703) may inhibit elongation by allosteric modification of the enzyme46.
Rifamycins were first isolated47 from the Gram-positive bacterium, Amycolatopsis mediterranei (originally Streptomyces mediterranei) in the 1950’s, and mutagenesis of this organism has led to the isolation and characterization of more potent rifamycin forms 48, including the clinically relevant rifamycin SV and rifampicin. In general, rifamycins are considered bactericidal against Gram-positive bacteria and bacteriostatic against Gram-negative bacteria, a difference that has been attributed to drug uptake and not β subunit affinity 49. Notably, rifamycins are among the first-line therapies used against Mycobacteria tuberculosis due to their efficient induction of cell death in mycobacterial species 50, although rifamycins are often used in combinatorial therapies owing to the rapid nature of resistance development49, 51.
Interestingly, an interaction between DNA and the hydroquinone moiety of RNA polymerase-bound rifamycin has been observed 52, and this interaction has been attributed to the location of the rifamycin drug molecule in relation to DNA in the DNA-RNA polymerase complex 42. This proximity, coupled with the reported ability of rifamycin to cycle between a radical and non-radical form (rifamycin SV and rifamycin S52, 53), may damage DNA through a direct drug-DNA interaction. This hypothesis may account for the observation that rifamycin SV can induce the SOS DNA damage response in E. coli, and that treatment of recA E. coli results in cell death whereas treatment of wildtype E. coli leads to bacteriostasis8.
Inhibition of cell wall synthesis
Lytic cell death
The bacterial cell is encased by layers of peptidoglycan (PG, or murein), a covalently cross-linked polymer matrix composed of peptide-linked β-(1–4)-N-acetyl hexosamine 54. The mechanical strength afforded by this layer of the cell wall is critical to a bacterium’s ability to survive environmental conditions that may alter prevailing osmotic pressures; of note, the degree of PG cross-linking can be correlated with the structural integrity of the cell 55. Maintenance of the PG layer is accomplished by the activity of transglycosylase and transpeptidase enzymes, which add disaccharide pentapeptides to extend the glycan strands of existing PG molecules and cross-link adjacent peptide strands of immature PG units, respectively56.
β-lactams and glycopeptides are among the classes of antibiotics that interfere with specific steps in homeostatic cell wall biosynthesis. Successful treatment with a cell wall synthesis inhibitor can result in changes to cell shape and size, induce cellular stress responses, and culminate in cell lysis6 (Figure 1b). For example, β-lactams (including penicillins, carbapenems and cephalosporins) block the cross-linking of PG units by inhibiting the peptide bond formation reaction catalyzed by transpeptidases, which are also known as penicillin-binding proteins (PBP)55, 57, 58. This inhibition is achieved by penicilloylation of a PBP’s transpeptidase active site –- the β-lactam drug molecule (containing a cyclic amide ring) is an analog of the terminal D-alanyl-D-alanine dipeptide of PG, and acts a substrate for the enzyme during the acylation phase of cross-link formation –- which disables the enzyme due to its inability to hydrolyze the bond created with the now ring-opened drug59, 60.
By contrast, most actinobacteria-derived glycopeptide antibiotics (e.g., vancomycin) inhibit PG synthesis through binding with PG units (at the D-alanyl-D-alanine dipeptide) and by blocking transglycosylase and transpeptidase activity 61. In this manner, glycopeptides (whether free in the periplasm like vancomycin or membrane-anchored like teicoplanin 62) generally act as steric inhibitors of PG maturation and reduce cellular mechanical strength, although some chemically-modified glycopeptides have been shown to directly interact with the transglycosylase enzyme 63. It is worth noting that β-lactams can be used to treat Gram-positive and Gram-negative bacteria, whereas glycopeptides are effective only against Gram-positive bacteria due to low permeability (Table 1). Additionally, antibiotics that inhibit the synthesis (e.g., Fosfomycin) and transport (e.g., Bacitracin) of individual PG units are also currently in use, as are lipopeptides (e.g., daptomycin) which affect structural integrity via their ability to insert into the cell membrane and induce depolarization.
Research into the mechanism of killing by PG synthesis inhibitors has centered on the lysis event. Initially, it was thought that inhibition of cell wall synthesis by β-lactams caused cell death when “internal pressure” built up due to growth outpacing cell wall expansion, resulting in lysis6. This “unbalanced growth” hypothesis was based in part on the notion that active protein synthesis is required for lysis to occur following the addition of β-lactams.
The lysis-dependent cell death mechanism, however, has proven to be much more complex, involving many active cellular processes. Seminal work demonstrated that Streptococcus pneumoniae deficient in amidase activity (murein hydrolase or autolysin activity) neither grew nor died following treatment with a lysis-inducing concentration of a β-lactam, an effect known as antibiotic tolerance 64. Autolysins are membrane-associated enzymes that break down bonds between and within PG strands, making them important during normal cell wall turnover and maintenance of cell shape 55. Autolysins have also been demonstrated to play a role in lytic cell death in bacterial species that contain numerous murein hydrolases, such as E. coli 65. In E. coli, a set of putative peptidoglycan hydrolases (LytM-domain factors) were shown to be important for rapid ampicillin-mediated lysis 66. The discovery that autolysins contribute to cell death expanded our understanding of lysis and showed that active degradation of the peptidoglycan layer by murein hydrolases, in conjunction with inhibition of peptidoglycan synthesis by a β-lactam antibiotic, will trigger lysis 64 (Figure 1b).
Non-lytic cell death
S. pneumoniae lacking murein hydrolase activity can still be killed by β-lactams, but at a slower rate than autolysin-active cells, indicating that there is a lysis-independent mode of killing induced by β-lactams64, 67. Evidence suggests that some of these non-lytic pathways are regulated by bacterial two-component systems 68. For example, in S. pneumoniae, the VncSR two-component system controls expression of the autolysin LytA, and regulates tolerance to vancomycin and penicillin through lysis-dependent 69 and lysis-independent 70 cell death pathways.
In Staphylococcus aureus, the LytSR two-component system can likewise affect cell lysis by regulating autolysin activity 71. LytR activates expression of lrgAB72, which was found to inhibit autolysin activity leading to antibiotic tolerance 73. LrgA is similar to bacteriophage holin proteins 73, which regulate the access of autolysins to the peptidoglycan layer. Based on this information, an additional holin-like system, cidAB, was uncovered in S. aureus and found to activate autolysins, rendering S. aureus more susceptible to β-lactam-mediated killing74, 75. Complementation of cidA into a cidA null strain was able to reverse the loss of autolysin activity, but it did not completely restore sensitivity to β-lactams 74.
Role of the SOS response in cell death by β-lactams
Treatment with β-lactams leads to changes in cell morphology that are associated with the primary drug-PBP interaction. Generally speaking, PBP1 inhibitors cause cell elongation and are potent triggers of lysis, PBP2 inhibitors alter cell shape but do not cause lysis, while PBP3 inhibitors influence cell division and can induce filamentation 76. Interestingly, β-lactam sub-types have distinct affinities for certain PBPs, which correlate with the ability of these drugs to stimulate autolysin activity and induce lysis76, 77. Accordingly, PBP1-binding β-lactams are also the most effective inducers of murein hydrolase activity and PBP2 inhibitors are the least proficient autolysin activators 77.
Filamentation can occur, following activation of the DNA damage responsive SOS network of genes 78 owing to expression of SulA, a key component of the SOS network which inhibits septation and leads to cell elongation by binding to and inhibiting polymerization of septation-triggering FtsZ monomers79, 80. Interestingly, β-lactams that inhibit PBP3 and induce filamentation have been shown to stimulate the DpiAB two-component system, which can activate the SOS response 81. β-lactam lethality can be enhanced by disrupting DpiAB signaling or a knocking-out the sulA gene, indicating that SulA activity may protect against a facet of β-lactam killing by shielding FtsZ and limiting a division ring interaction among PBPs and murein hydrolases. In support of this idea, sulA expression limits the lysis observed in a strain of E. coli expressing FtsZ84 (a mutant of FtsZ active only under certain temperature and media conditions) and lacking PBP4 and PBP782.
DNA damaging antimicrobials, such as quinolones that do not directly disrupt murein turnover, also cause filamentation through induction of the SOS response4. Interestingly, a mutant strain of E. coli (W7) deficient in diaminopimelic acid synthesis, a key building block of PG, will undergo lysis following treatment with the fluoroquinolone antimicrobials ofloxacin or pefloxacin 83. This finding points toward roles for PG turnover and the SOS response in antibiotic-mediated lytic killing responses.
Inhibition of protein synthesis
The process of mRNA translation occurs over three sequential phases (initiation, elongation and termination) involving the ribosome and a host of cytoplasmic accessory factors 84. The ribosome organelle is composed of two ribonucleoprotein subunits, the 50S and 30S, which organize (initiation phase) on the formation of a complex between an mRNA transcript, fMet-charged aminoacyl-tRNA, several initiation factors and a free 30S subunit 85. Drugs that inhibit protein synthesis are among the broadest classes of antibiotics and can be divided into two subclasses: the 50S inhibitors and 30S inhibitors (Table 1).
50S ribosome inhibitors include the macrolide (e.g., erythromycin), lincosamide (e.g., clindamycin), streptogramin (e.g., dalfopristin/quinupristin), amphenicol (e.g., chloramphenicol) and oxazolidinone (e.g., linezolid) classes of antibiotics86, 87. In general terms, 50S ribosome inhibitors work by physically blocking either initiation of protein translation (as is the case for oxazolidinones 88), or translocation of peptidyl-tRNAs, which serves to inhibit the peptidyltransferase reaction that elongates the nacent peptide chain. Studies of macrolide, lincosamide and streptogramin drugs have provided for a mode-of-action model that involves blocking the access of peptidyl-tRNAs to the ribosome (to varying degrees), subsequent blockage of the peptidyltransferase elongation reaction by steric inhibition, and eventually triggering dissociation of the peptidyl-tRNA89, 90. This model also accounts for the phenomenon that these classes of drugs lose their antibacterial activity when elongation has progressed beyond a critical length91.
30S ribosome inhibitors include the tetracycline and aminocyclitol families of antibiotics. Tetracyclines work by blocking the access of aminoacyl-tRNAs to the ribosome92. The aminocyclitol class is comprised of spectinomycin and the aminoglycoside family of antibiotics (for example, streptomycin, kanamycin and gentamicin), which bind the 16S rRNA component of the 30S ribosome subunit. Spectinomycin, interferes with the stability of peptidyl-tRNA binding to the ribosome by inhibiting elongation factor-catalyzed translocation, but does not cause protein mistranslation 93–95. By contrast, the interaction between aminoglycosides and the 16S rRNA can induce an alteration in the conformation of the complex formed between an mRNA codon and its cognate charged aminoacyl-tRNA at the ribosome, promoting tRNA mismatching which can result in protein mistranslation96–99.
Among ribosome inhibitors, the naturally-derived aminoglycoside subclass is the only one that is broadly bactericidal. Macrolides, streptogramins, spectinomycin, tetracyclines, chloramphenicol and macrolides are typically bacteriostatic, however, these families of ribosome inhibitors can be bactericidal in a species- or treatment-specific fashion. For example, chloramphenicol has been shown to kill S. pneumoniae and Neisseria meningitides effectively 100, while chloramphenicol and the macrolide, azythromycin, have exhibited bactericidality against Haemophilus influenza100, 101. This species-specific variability in ribosome inhibitor-mediated cell death likely has to do with sequence differences among bacterial species in the variable regions of the highly conserved ribosomal proteins and RNAs 102. Additionally, high concentrations of macrolides and combinations of streptogramin group A and B drug molecules are capable of behaving bactericidal-like. For the remainder of this section, however, we will focus on the aminoglycoside class of drugs, for which the mechanism of killing by ribosome inhibition has best been studied.
Aminoglycoside uptake and cell death
Binding of aminoglycosides to the ribosome does not bring translation to an immediate standstill. Rather, as noted above, this class of drugs promotes protein mistranslation through the incorporation of inappropriate amino acids into elongating peptide strands 96, a phenotype specific for aminoglycosides and one which contributes to cell killing (Figure 1c).
Respiration also plays a crucial role in aminoglycoside uptake and lethality 103. Following the initial step of drug molecule adsorption (in Gram-negative models like E. coli) through electrostatic interaction, changes in membrane potential provide for cellular access by aminoglycosides. Respiration-dependent uptake relies on the activity of membrane-associated cytochromes and maintenance of the electrochemical potential through the quinone pool104, 105. Accordingly, under anaerobic conditions, aminoglycoside uptake is severely limited in both Gram-positive and Gram-negative bacteria106, 107, although there is evidence that aminoglycoside uptake can occur under certain anaerobic conditions via a mechanism that is sensitive to nitrate levels. In E. coli and P. aeruginosa, aminoglycoside uptake can take place when nitrate is used as an electron acceptor in place of oxygen, and anaerobic bacteria that have quinones and cytochromes can take up aminoglycosides if significant anaerobic electron transport occurs 108.
Additionally, aminoglycoside-mediated killing has been linked in E. coli with alterations to the cell membrane ultrastructure that ultimately increase drug uptake109, 110. Aminoglycosides can affect membrane composition through incorporation of mistranslated membrane proteins into the cytoplasmic membrane, thereby increasing cellular permeability and affording for increased access of the drug molecule 103 (Figure 1c). Sufficient aminoglycoside uptake resulting in increased ribosome inhibition and cell death could also occur as a function of the changes in membrane integrity owing to the incorporation of mistranslated membrane proteins 103. An alteration in membrane permeability due to aminoglycoside-induced membrane damage is believed to be one of the mechanisms whereby aminoglycosides are synergistic with β-lactams (see Box 1 for more on drug synergy and antagonism).
Box 1 . Drug synergy.
Combinatorial antibiotic treatments can have diverse effects on bacterial survival. Antibiotics can be more effective as a combination treatment displaying either an additive effect (effect equal to sum of treatments) or a synergistic effect (effect greater than sum of treatments); the combination can also be antagonistic, i.e., the effect of the combination treatment is less than the effect of the respective single-drug treatments 136. Technological advances have allowed for high-throughput quantification of drug-drug interactions at the level of cell survival and target binding, thereby opening the door for the systematic study of synergistic and antagonistic combinations137.
The exploration of the survival fitness landscape between drug combinations has allowed for the study of the mechanisms by which antibiotics work against bacteria 138 and has also allowed for a study of the evolution of drug resistance 137. Further study of the synergy or antagonism between antibiotics will provide additional insight into the underlying cell death mechanisms for the individual classes of antibiotics. For example, the suppressive interaction between protein synthesis inhibitors and DNA synthesis inhibitors has been shown to be due to non-optimal ribosomal RNA regulation by DNA-inhibiting drugs139.
The synergy between aminoglycosides and β-lactams has been attributed to β-lactam-mediated membrane damage leading to increased uptake of aminoglycosides 140. It will be interesting to see if the synergy between these two drugs is also related to the induction of the envelope stress response that has been observed with aminoglycosides10.
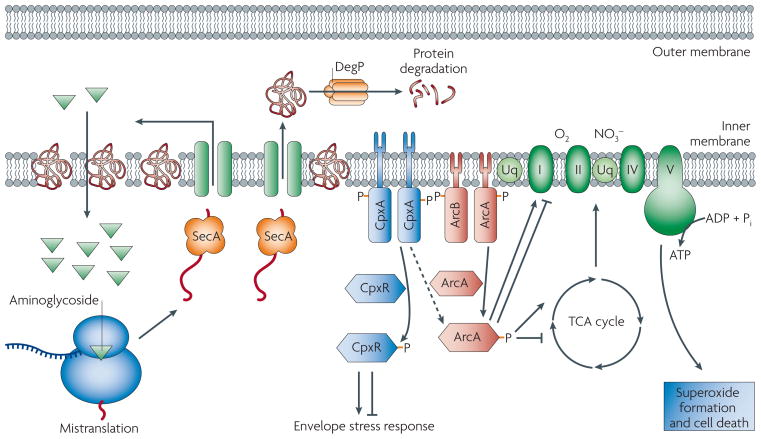
Another consequence of mistranslated protein incorporation into the bacterial membrane is the activation of envelope (Cpx) and redox-responsive (Arc) two-component systems. These intracellular signal relay systems regulate the expression of genes important for the maintenance of membrane integrity and composition 111, and membrane-coupled energy generation112, 113, respectively. Disruption of Cpx or Arc two-component system signaling (through a series of single-gene knockouts) has recently been shown to reduce the killing efficacy of aminoglycosides, a result associated with findings linking bactericidal antibiotic-induced cell death with drug stress-induced changes in metabolism. Interestingly, disruption of Cpx or Arc two-component system signaling was also shown to reduce the lethality of β-lactam and quinolone antibiotics10. Together, these findings point toward a broad role for the envelope stress responsive and redox responsive two-component systems in killing by bactericidal drugs (Figure 2).
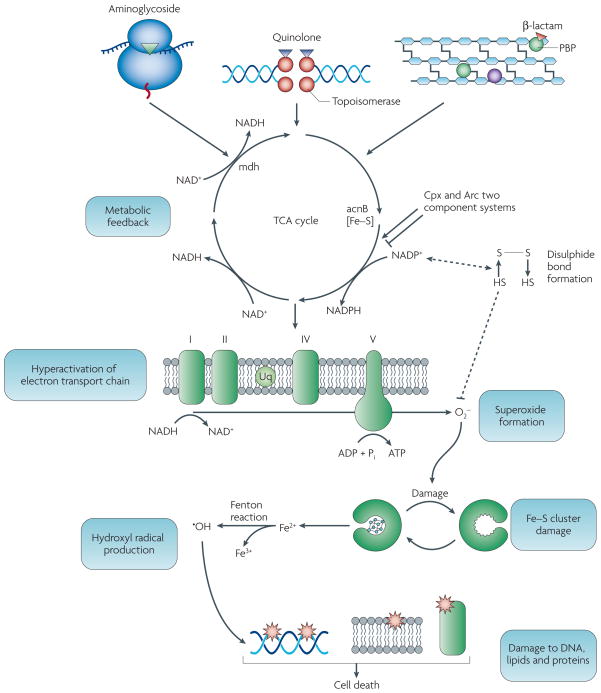
Figure 2. Common mechanism of cell death induced by bactericidal antibiotics.
The primary drug-target interactions (aminoglycoside with the ribosome, quinolone with DNA gyrase, and β-lactam with penicillin-binding proteins) stimulate oxidation of NADH through the electron transport chain that is dependent on the TCA cycle. Hyperactivation of the electron transport chain stimulates superoxide formation. Superoxide damages iron-sulfur clusters, making ferrous iron available for oxidation by the Fenton reaction. The Fenton reaction leads to hydroxyl radical formation and the hydroxyl radicals damage DNA, proteins and lipids, which contributes to antibiotic-induced cell death. Quinolones, β-lactams and aminoglycosides also trigger radical formation and cell death through the Cpx and Arc two-component systems. It is also possible that redox-sensitive proteins such as those containing disulfide contribute in an as yet undetermined fashion to the common mechanism (dashed lines). (Modified with permission from ref 8)
Antibiotic network biology
As noted above, antibiotic-mediated cell death is a complex process that only begins with the drug-target interaction and the primary effects of these respective interactions. Looking forward, the development of novel antibiotics and improvement of current antibacterial drug therapies would benefit from a better understanding of the specific sequences of events beginning with the binding of a bactericidal drug molecule to its target, and ending in cell death.
Bioinformatics approaches that utilize high-throughput genetic screening or gene expression profiling have proven to be valuable tools to explore the response layers of bacteria to varying antibiotic treatments 114. For example, recent screens for antibiotic susceptibility in a single-gene deletion library of non-essential genes in E. coli 115 and a transposon mutagenesis library in Pseudomonas aeruginosa 116 have provided important insights into the numbers and types of genes that affect treatment efficiency (bactericidal versus bacteriostatic effects), including those related to drug molecule efflux, uptake, or degradation. Additionally, monitoring of global changes in gene expression patterns, or signatures, resulting from antibiotic treatment over a range of conditions has advanced our understanding of the off target effects elicited by primary drug-target interactions 114.
A need also exists for the application of network biology methods to discern and resolve the potential interplay between genes and proteins coordinating bacterial stress response pathways. Typically, such methods incorporate gene expression profiling data and the results of high-throughput genetic screens, along with the contents of databases detailing experimentally identified regulatory connections and biochemical pathway classifications, to functionally enrich datasets and predict relationships that exist among genes under tested conditions. As such, biological network studies of drug-treated bacteria can be used to advance our understanding of how groups of genes interact functionally, rather than in isolation, when cells react to antibiotic stress 117.
To help address this problem, researchers have developed methods to construct quantitative models of regulatory networks 118–122, and have recently used these reconstructed network models to identify the sets of genes, associated functional groups and biochemical pathways that act in concert to mediate bacterial responses to antibiotics8–10, 119. Below we highlight some mechanistic insights that have been gleaned from antibiotic network biology, and discuss some opportunities and challenges for this emerging area of research.
A common mechanism for antibiotic-mediated cell death
As an example of the utility of studying bacterial stress responses at the systems level, biological network analysis methods were recently employed to identify novel mechanisms that contribute to bacterial cell death upon DNA gyrase inhibition by the fluoroquinolone antibiotic, norfloxacin9. As noted above, quinolones are known to induce cell death through the introduction of double-stranded DNA breaks following arrest of topoisomerase function4. To identify additional cellular contributions to cell death resulting from gyrase poisoning, reconstruction of stress response networks was performed following treatment of E. coli with lethal concentrations of norfloxacin. In the course of this work, a novel oxidative damage cell death pathway, which involves reactive oxygen species generation and a breakdown in iron regulatory dynamics following norfloxacin-induced DNA damage induction, was uncovered. More specifically, norfloxacin treatment was found to promote superoxide generation soon after gyrase poisoning, and was ultimately shown to result in the generation of highly-destructive hydroxyl radicals through the Fenton reaction 123. Under these conditions, the Fenton reaction was found to be fueled by superoxide-mediated destabilization of iron-sulfur cluster catalytic sites, repair of these damaged iron-sulfur clusters, and related changes in iron-related gene expression9
Building on this work, it was later shown that all major classes of bactericidal antibiotics (β-lactams, aminoglycosides, quinolones), despite the stark differences in their primary drug-target interactions, promote the generation of lethal hydroxyl radicals in both Gram-negative and Gram-positive bacteria8. Stress response network analysis methods employed in this study suggested that antibiotic-induced hydroxyl radical formation is the end product of a common mechanism, wherein alterations in central metabolism related to NADH consumption (increased TCA cycle and respiratory activity) are critical to superoxide-mediated iron-sulfur cluster destabilization and stimulation of the Fenton reaction. These predictions were validated by the results of additional phenotypic experiments, biochemical assays and gene expression measurements, confirming that lethal levels of bactericidal antibacterials trigger a common oxidative damage cellular death pathway, which contributes to killing by these drugs (Figure 2).
Most recently, the study of antibiotic-induced stress response networks has been aimed at determining exactly how the primary effect of a given bactericidal drug triggers aspects of cell death that are common to all bactericidal drugs. For example, a comparative analysis of stress response networks, reconstructed using gene expression data from aminocyclitol-treated (spectinomycin, gentamicin and kanamycin) E. coli, was used to identify the incorporation of mistranslated proteins into the cell membrane as the trigger for aminoglycoside-induced oxidative stress (Figure 3)10. Interestingly, mistranslated membrane proteins were shown to stimulate radical formation by activating the envelope stress (Cpx) and redox-responsive (Arc) two-component systems, ultimately altering TCA cycle metabolism; the TCA cycle had previously been implicated in aminoglycoside susceptibility8, 124.
Figure 3. Aminoglycoside triggers for radical-mediated cell death.
The interaction between the aminglycoside and ribosome causes mistranslation and misfolding of membrane proteins. Incorporation of mistranslated, misfolded proteins into the cell membrane stimulates the envelope (Cpx) and redox-responsive (Arc) two-component systems. Activation of these systems perturbs cellular metabolism and the membrane potential (DY), resulting in the formation of lethal hydroxyl radicals. (Modified with permission from ref 10)
The discovery of the common oxidative damage cellular death pathway has important implications for the development of more effective antibacterial therapies. Specifically, it indicates that all major classes of bactericidal drugs can be potentiated by inhibition of the DNA stress response network (i.e., the SOS response), which plays a key part in the remediation of hydroxyl radical-induced DNA damage. This may be accomplished through the development of small molecules (e.g., RecA inhibitors 125) or synthetic biology approaches (see Box 2).
Box 2. Synthetic biology for antibacterial applications.
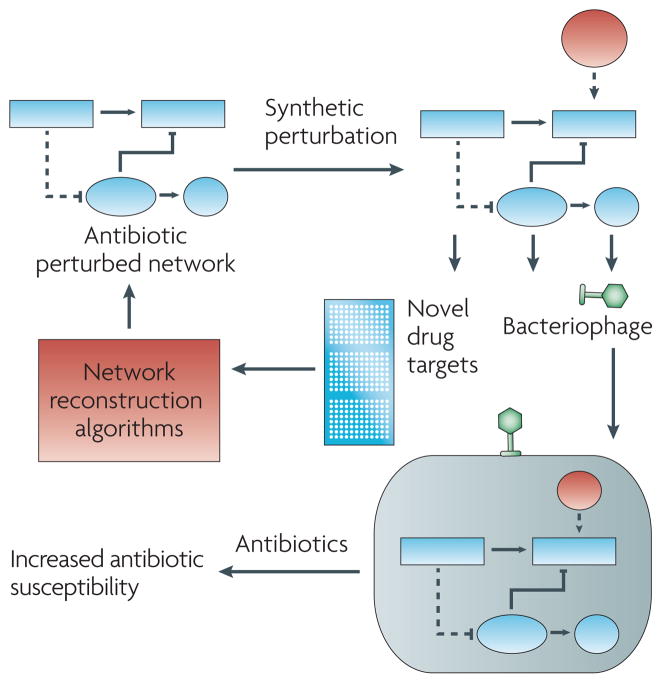
The study of complex antibiotic-related cell death systems can also be aided by synthetic biology. Delivery of engineered gene circuits that alter response network behavior can serve as a tool to experimentally interrogate antibiotic-mediated cell death pathways, as well as a means to enhance killing by an antibiotic (Figure 4). Bacteriophage, which are bacteria-specific viruses, show promise as an effective means to deliver “network perturbations” to bacteria so as to improve antibiotic lethality141, 142. Bacteriophage have been used to enhance killing of E. coli by bactericidal antibiotics through the delivery of proteins that modify the oxidative stress response or inhibit DNA damage repair systems 142. Bacteriophage are species specific, so it may be possible to use engineered bacteriophage to deliver antibiotic-enhancing synthetic gene networks, therapeutic proteins or antimicrobial peptides that are highly specific for an infecting organism. This would allow for efficient treatment of a bacterial infection, while sparing the typical commensal body flora (Figure 4).
Box 2 inset. Network biology approaches for antibiotic functional analysis and therapeutic development.
Introduction of novel connections or alteration of the connectivity among genes in an antibiotic-related network can be used to refine or expand our knowledge of these networks and interrogate drug networks for novel antibiotic targets. Synthetic gene networks, therapeutic proteins or antimicrobial peptides that enhance antibiotic efficacy can also be delivered directly to bacteria through species-specific delivery mechanisms such as bacteriophage.
Reactive oxygen species, such as superoxide and hydroxyl radicals, are highly toxic and have deleterious effects on bacterial physiology123, 126, 127, even under steady-state conditions. There is still much to be learned about how oxidative stress-related changes in bacterial physiology affect bactericidal antibiotic-mediated cell death and the emergence of resistance128, 129. For example, it was recently discovered that endogenous nitric oxide (NO) produced by bacteria with NO synthases can protect against ROS-mediated cell death 130. Additionally, considering bacteria have developed mechanisms to avoid phagocyte-generated ROS in the immune response to infection 131, it will be interesting to explore, from a systems-level perspective, the relationship between immune-mediated cell death and drug-mediated cell death.
Opportunities and challenges for antibiotic network biology
One of the more intriguing aspects of antibacterial therapies is that not all bacterial species respond in the same way to antibiotic treatment. Network biology approaches, which provide the field of antibiotic research with an opportunity to view response mechanisms of different bacterial species to various classes of antibiotics, could be extended to the context of particular infectious species, persistent infections or disease settings. As an example, it is generally accepted that Gram-negative bacteria are not susceptible to the glycopeptide, vancomycin, or the depolarizing lipopeptide, daptomycin, yet a single gene, yfgL, was recently found that can make E. coli susceptible to glycolipid derivatives of vancomycin 132. Gene expression profiling of daptomycin-treated S. aureus has revealed that daptomycin perturbs PG synthesis, through a mechanism involving activation of cell-wall stress systems and membrane depolarization 133. Given these findings, we may be able to combine our knowledge of β-lactam- and aminoglycoside-induced gene signatures, with the results of high-throughput screens at various drug doses, to reconstruct drug-specific cell death networks that use YgfL as a “network anchor”. Predicted functional and regulatory relationships between enriched genes could then be used to determine the secondary effects of lipopeptide antibiotics and gain insight into the differences in killing by this drug in Gram-negative and Gram-positive bacteria.
Moreover, the development of comparative network biology techniques will be essential to developing our understanding of how species-specific differences manifest themselves in divergent drug-specific cell death networks and variations in physiological responses. These methods may be particularly useful when examining pathogenic bacteria with sparse systems-level data (such as Shigella or Salmonella species) that are closely related to well-studied bacteria (such as E. coli). Through a greater understanding of the biological networks related to an individual drug target, we eventually might be able to search for meaningful network homologues among species in the same spirit as one currently searches for gene homologues. Network-based efforts may also lead to the development of species-specific treatments, including synthetic biology-derived therapies (see Box 2) which could be useful in killing off harmful, invasive bacteria, while leaving our normal bacterial flora intact.
Finally, bacterial network analyses will also be useful in the study of non-classical antibacterial agents that induce cell death. Antimicrobial peptides (AMPs) are short cationic peptides which are thought to kill through interactions with the membrane that result in pore formation134, 135. However, the mode of action of many AMPs may, in fact, be more complex, and cell death networks uncovered for existing antibiotics could be used as mechanistic templates to study cellular responses induced by AMPs.
Concluding remarks
Drug-resistant bacterial infections are becoming more prevalent and are a major health issue facing us today. This rise in resistance has limited our repertoire of effective antimicrobials, creating a problematic situation which has been exacerbated by the small number of new antibiotics introduced in recent years. The complex effects of bactericidal antibiotics discussed in this review provide a large playing field for the development of novel antibacterial compounds, as well as adjuvant molecules and synthetic biology constructs that could enhance the potency of current antibiotics. It will be important to translate our growing understanding of antibiotic mechanisms into new clinical treatments and approaches, so that we can effectively fight the growing threat from resistant pathogens.
Acknowledgments
We thank the anonymous Reviewers for their helpful comments and suggestions. This work was supported by the National Institutes of Health through the NIH Director’s Pioneer Award Program, grant number DP1 OD003644, and the Howard Hughes Medical Institute.
Glossary Terms
- Antibiotic tolerance
Bacteria that neither grow nor die following exposure to lethal concentrations of bactericidal antibiotics
- Antimicrobial peptides
Short, naturally occurring cationic peptides that have antibacterial properties through their ability to interfere with bacterial membranes
- Autolysins
Enzymes that hydrolyze the β-linkage between the monosaccharide monomers in peptidoglycan units, and can induce lysis when in excess
- Bacteriostatic
Antibiotics that inhibit growth with no loss of viability
- Bactericidal
Antibiotics that inhibit growth and kill bacteria upon exposure
- Cell envelope
Layers of the cell surrounding the cytoplasm which include lipid membranes and peptidoglycan layers
- Fenton reaction
Reaction of ferrous iron (FeII) with hydrogen peroxide to produce ferric iron (FeIII) and a hydroxyl radical
- Free radicals
Molecules containing an unpaired electron
- Lysis
Rupture of the cell envelope leading to the expulsion of intracellular contents into the surrounding environment with eventual disintegration of the cell envelope
- Murein hydrolases
Enzymes that introduce cuts, between carbon-nitrogen non-peptide bonds, while pruning the peptidoglycan layer and are important for homeostatic murein turnover
- Gram-positive bacteria
Bacterial species whose outer peptidoglycan envelope layer is stained blue/violet by crystal violet during Gram staining
- Gram-negative bacteria
Bacterial species whose lipopolysaccharide-containing outer membrane (surrounding the periplasmic and peptidoglycan envelope layers) can exclude crystal violet and are instead stained red/pink by a counterstain during Gram staining
- Oxidative phosphorylation
Process of ATP generation driven by the electrochemical gradient maintained by the electron transport chain
- Peptidoglycan
A mesh-like matrix of covalently cross-linked polymers, composed of peptide bond-linked β-(1–4)-N-acetyl hexosamine, that compose the cell envelope or murein layer
- Persisters
A subpopulation of bacteria that survive the initial rapid cell death observed following treatment with a bactericidal antibiotic that do not actively grow in the presence of the bactericidal antibiotic, but will repopulate a culture following removal of the antibiotic
- SOS response
The DNA stress response pathway in E. coli, whose prototypical network of genes is regulated by the transcriptional repressor, LexA, and is commonly activated by the co-regulatory protein RecA, which promotes LexA autocleavage when activated; a growing number of studies have explored the intriguing dynamics of this network, and alternative mechanisms of activation.
- Toxin-antitoxin systems
Genetic modules that consist of a “toxin” protein which has a detrimental effect on cell growth or survival, and a cognate “antitoxin” that protects against toxin activity by sequestration of the toxin
- Two-component system
A two-protein signal-relay system composed of a sensor histidine kinase and a cognate receiver protein, which is typically a transcription factor
- Quinone pool
Membrane-associated cyclic aromatic-based compounds that shuttle electrons along the electron transport chain
Biographies
Michael Kohanski: Michael Kohanski received a B.S. in bioengineering from the University of Pennsylvania and a Ph.D. in biomedical engineering from Boston University in the lab of James Collins. He is currently working toward the completion of his M.D. from Boston University School of Medicine. His research focuses on using systems and network biology approaches to gain new insights into antibiotic-mediated cell death. He is interested in applying these approaches to better understand and explore the host-microbe interaction and to develop novel antibacterial therapies.
Daniel Dwyer: Daniel Dwyer is an alumnus of Boston College (B.S. in Biology, with Erik Flemington at the Dana-Farber Cancer Institute) and Boston University (Ph.D. in Molecular Biology, Cell Biology and Biochemistry, with James Collins); he has also received training at New York University Medical School (with Miguel Gama-Sosa) and in the biotechnology industry at Forticell Bioscience (with Lee McDonald and Stephanie Knight). He is currently an NIH post-doctoral fellow in the Department of Biomedical Engineering at Boston University. He is using a variety of approaches and tools to characterize the broad biochemical responses of bacteria to antibacterial treatments, and the contribution of these elicited phenotypes to cell death.
James Collins: James J. Collins is an Investigator of the Howard Hughes Medical Institute, and a William F. Warren Distinguished Profesor, University Professor, Professor of Biomedical Engineering, and Co-Director of the Center for BioDynamics at Boston University. He is also a core founding faculty member of the Wyss Institute for Biologically Inspired Engineering at Harvard University. His research group works in synthetic biology and systems biology, with a particular focus on network biology approaches to antibiotic action and bacterial defense mechanisms.
References
- 1.Walsh C. Antibiotics: actions, origins, resistance. ASM Press; Washington, D.C: 2003. [Google Scholar]
- 2.Fleming A. On antibacterial action of culture of penicillium, with special reference to their use in isolation of B. influenzae. British Journal of Experimental Pathology. 1929;10:226–236. [PubMed] [Google Scholar]
- 3.Taubes G. The bacteria fight back. Science. 2008;321:356–61. doi: 10.1126/science.321.5887.356. [DOI] [PubMed] [Google Scholar]
- 4.Drlica K, Malik M, Kerns RJ, Zhao X. Quinolone-mediated bacterial death. Antimicrob Agents Chemother. 2008;52:385–92. doi: 10.1128/AAC.01617-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Floss HG, Yu TW. Rifamycin-mode of action, resistance, and biosynthesis. Chem Rev. 2005;105:621–32. doi: 10.1021/cr030112j. [DOI] [PubMed] [Google Scholar]
- 6•.Tomasz A. The mechanism of the irreversible antimicrobial effects of penicillins: how the beta-lactam antibiotics kill and lyse bacteria. Annu Rev Microbiol. 1979;33:113–37. doi: 10.1146/annurev.mi.33.100179.000553. This seminal review of beta-lactam-mediated cell death discusses the intricacies of killing by various members of this antibiotic class in terms of the specific drug-inhibited protein targets and their related cell wall maintenance functions. [DOI] [PubMed] [Google Scholar]
- 7.Vakulenko SB, Mobashery S. Versatility of aminoglycosides and prospects for their future. Clin Microbiol Rev. 2003;16:430–50. doi: 10.1128/CMR.16.3.430-450.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8•.Kohanski MA, Dwyer DJ, Hayete B, Lawrence CA, Collins JJ. A common mechanism of cellular death induced by bactericidal antibiotics. Cell. 2007;130:797–810. doi: 10.1016/j.cell.2007.06.049. Reveals that treatment of Gram-positive and Gram-negative bacteria with lethal levels of bactericidal antibiotics induces the formation of hydroxyl radicals via a common mechanism involving drug-induced changes in NADH consumption and central metabolism, notably the tricarboxylic acid cycle. [DOI] [PubMed] [Google Scholar]
- 9•.Dwyer DJ, Kohanski MA, Hayete B, Collins JJ. Gyrase inhibitors induce an oxidative damage cellular death pathway in Escherichia coli. Mol Syst Biol. 2007;3:91. doi: 10.1038/msb4100135. Describes the physiological responses of E. coli following inhibition of DNA gyrase by a fluoroquinolone antibiotic and a peptide toxin, include activation of the superoxide stress response and increased iron-sulfur cluster synthesis. Ultimately, these physiological changes are shown to result in hydroxyl radical production, which contribute to cell death. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10•.Kohanski MA, Dwyer DJ, Wierzbowski J, Cottarel G, Collins JJ. Mistranslation of membrane proteins and two-component system activation trigger antibiotic-mediated cell death. Cell. 2008;135:679–90. doi: 10.1016/j.cell.2008.09.038. Demonstrates that systems which facilitate membrane protein traffic are central to aminoglycoside-induced oxidative stress and cell death. This occurs by signaling through the redox- and the envelope stress-responsive two-component systems. These two-component systems are also shown to have a general role in bactericidal antibiotic-mediated oxidative stress and cell death, expanding our understanding of the common mechanism of killing induced by bactericidal antibiotics. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Espeli O, Marians KJ. Untangling intracellular DNA topology. Mol Microbiol. 2004;52:925–31. doi: 10.1111/j.1365-2958.2004.04047.x. [DOI] [PubMed] [Google Scholar]
- 12.Drlica K, Snyder M. Superhelical Escherichia coli DNA: relaxation by coumermycin. J Mol Biol. 1978;120:145–54. doi: 10.1016/0022-2836(78)90061-x. [DOI] [PubMed] [Google Scholar]
- 13.Gellert M, Mizuuchi K, O’Dea MH, Nash HA. DNA gyrase: an enzyme that introduces superhelical turns into DNA. Proc Natl Acad Sci U S A. 1976;73:3872–6. doi: 10.1073/pnas.73.11.3872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14•.Sugino A, Peebles CL, Kreuzer KN, Cozzarelli NR. Mechanism of action of nalidixic acid: purification of Escherichia coli nalA gene product and its relationship to DNA gyrase and a novel nicking-closing enzyme. Proc Natl Acad Sci U S A. 1977;74:4767–71. doi: 10.1073/pnas.74.11.4767. References 14 and 15 discuss the results of complementary in vivo and in vitro studies that characterized the genetic locus (nalA, later gyrA) and the basic mechanism of quinolone antibiotic action (prevention of DNA duplex strand rejoining yielding double-stranded DNA breaks), while postulating on the composition and energetic requirements of DNA gyrase activity. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15•.Gellert M, Mizuuchi K, O’Dea MH, Itoh T, Tomizawa JI. Nalidixic acid resistance: a second genetic character involved in DNA gyrase activity. Proc Natl Acad Sci U S A. 1977;74:4772–6. doi: 10.1073/pnas.74.11.4772. References 14 and 15 discuss the results of complementary in vivo and in vitro studies that characterized the genetic locus (nalA, later gyrA) and the basic mechanism of quinolone antibiotic action (prevention of DNA duplex strand rejoining yielding double-stranded DNA breaks), while postulating on the composition and energetic requirements of DNA gyrase activity. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hooper DC, Rubinstein E. Quinolone antimicrobial agents. ASM Press; Washington, D.C: 2003. [Google Scholar]
- 17.Rubinstein E. History of quinolones and their side effects. Chemotherapy. 2001;47(Suppl 3):3–8. doi: 10.1159/000057838. discussion 44–8. [DOI] [PubMed] [Google Scholar]
- 18.Lu T, et al. Enhancement of fluoroquinolone activity by C-8 halogen and methoxy moieties: action against a gyrase resistance mutant of Mycobacterium smegmatis and a gyrase-topoisomerase IV double mutant of Staphylococcus aureus. Antimicrob Agents Chemother. 2001;45:2703–9. doi: 10.1128/AAC.45.10.2703-2709.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19•.Chen CR, Malik M, Snyder M, Drlica K. DNA gyrase and topoisomerase IV on the bacterial chromosome: quinolone-induced DNA cleavage. J Mol Biol. 1996;258:627–37. doi: 10.1006/jmbi.1996.0274. Identifies topoisomerase IV as a second target of fluoroquinolone antibiotics in Gram-negative bacteria, while characterizing subtle yet critical differences in the mechanism of killing by various quinolone drugs. [DOI] [PubMed] [Google Scholar]
- 20.Drlica K, Zhao X. DNA gyrase, topoisomerase IV, and the 4-quinolones. Microbiol Mol Biol Rev. 1997;61:377–92. doi: 10.1128/mmbr.61.3.377-392.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Munoz R, De La Campa AG. ParC subunit of DNA topoisomerase IV of Streptococcus pneumoniae is a primary target of fluoroquinolones and cooperates with DNA gyrase A subunit in forming resistance phenotype. Antimicrob Agents Chemother. 1996;40:2252–7. doi: 10.1128/aac.40.10.2252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Belland RJ, Morrison SG, Ison C, Huang WM. Neisseria gonorrhoeae acquires mutations in analogous regions of gyrA and parC in fluoroquinolone-resistant isolates. Mol Microbiol. 1994;14:371–80. doi: 10.1111/j.1365-2958.1994.tb01297.x. [DOI] [PubMed] [Google Scholar]
- 23.Critchlow SE, Maxwell A. DNA cleavage is not required for the binding of quinolone drugs to the DNA gyrase-DNA complex. Biochemistry. 1996;35:7387–93. doi: 10.1021/bi9603175. [DOI] [PubMed] [Google Scholar]
- 24.Marians KJ, Hiasa H. Mechanism of quinolone action. A drug-induced structural perturbation of the DNA precedes strand cleavage by topoisomerase IV. J Biol Chem. 1997;272:9401–9. doi: 10.1074/jbc.272.14.9401. [DOI] [PubMed] [Google Scholar]
- 25•.Kampranis SC, Maxwell A. The DNA gyrase-quinolone complex. ATP hydrolysis and the mechanism of DNA cleavage. J Biol Chem. 1998;273:22615–26. doi: 10.1074/jbc.273.35.22615. Reveals that quinolone antibiotic binding to the gyrase-DNA complex occurs before DNA strand breakage and that DNA cleavage can occur, albeit at a slower rate, in the presence of the drug molecule based on the results of ATP hydrolysis and DNA cleavage assays. [DOI] [PubMed] [Google Scholar]
- 26.Yoshida H, Bogaki M, Nakamura M, Nakamura S. Quinolone resistance-determining region in the DNA gyrase gyrA gene of Escherichia coli. Antimicrob Agents Chemother. 1990;34:1271–2. doi: 10.1128/aac.34.6.1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Morais Cabral JH, et al. Crystal structure of the breakage-reunion domain of DNA gyrase. Nature. 1997;388:903–6. doi: 10.1038/42294. [DOI] [PubMed] [Google Scholar]
- 28.Heddle J, Maxwell A. Quinolone-binding pocket of DNA gyrase: role of GyrB. Antimicrob Agents Chemother. 2002;46:1805–15. doi: 10.1128/AAC.46.6.1805-1815.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Goss WA, Deitz WH, Cook TM. Mechanism of Action of Nalidixic Acid on Escherichia Coli. Ii. Inhibition of Deoxyribonucleic Acid Synthesis. J Bacteriol. 1965;89:1068–74. doi: 10.1128/jb.89.4.1068-1074.1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Snyder M, Drlica K. DNA gyrase on the bacterial chromosome: DNA cleavage induced by oxolinic acid. J Mol Biol. 1979;131:287–302. doi: 10.1016/0022-2836(79)90077-9. [DOI] [PubMed] [Google Scholar]
- 31.Cox MM, et al. The importance of repairing stalled replication forks. Nature. 2000;404:37–41. doi: 10.1038/35003501. [DOI] [PubMed] [Google Scholar]
- 32.Courcelle J, Hanawalt PC. RecA-dependent recovery of arrested DNA replication forks. Annu Rev Genet. 2003;37:611–46. doi: 10.1146/annurev.genet.37.110801.142616. [DOI] [PubMed] [Google Scholar]
- 33.Howard BM, Pinney RJ, Smith JT. Function of the SOS process in repair of DNA damage induced by modern 4-quinolones. J Pharm Pharmacol. 1993;45:658–62. doi: 10.1111/j.2042-7158.1993.tb05673.x. [DOI] [PubMed] [Google Scholar]
- 34.Cirz RT, et al. Inhibition of mutation and combating the evolution of antibiotic resistance. PLoS Biol. 2005;3:e176. doi: 10.1371/journal.pbio.0030176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Guerin E, et al. The SOS response controls integron recombination. Science. 2009;324:1034. doi: 10.1126/science.1172914. [DOI] [PubMed] [Google Scholar]
- 36.Beaber JW, Hochhut B, Waldor MK. SOS response promotes horizontal dissemination of antibiotic resistance genes. Nature. 2004;427:72–4. doi: 10.1038/nature02241. [DOI] [PubMed] [Google Scholar]
- 37.Lewin CS, Howard BM, Smith JT. Protein- and RNA-synthesis independent bactericidal activity of ciprofloxacin that involves the A subunit of DNA gyrase. J Med Microbiol. 1991;34:19–22. doi: 10.1099/00222615-34-1-19. [DOI] [PubMed] [Google Scholar]
- 38.Wang X, Zhao X, Malik M, Drlica K. Contribution of reactive oxygen species to pathways of quinolone-mediated bacterial cell death. J Antimicrob Chemother. 2010;65:520–4. doi: 10.1093/jac/dkp486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kolodkin-Gal I, Sat B, Keshet A, Engelberg-Kulka H. The communication factor EDF and the toxin-antitoxin module mazEF determine the mode of action of antibiotics. PLoS Biol. 2008;6:e319. doi: 10.1371/journal.pbio.0060319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dukan S, et al. Protein oxidation in response to increased transcriptional or translational errors. Proc Natl Acad Sci U S A. 2000;97:5746–9. doi: 10.1073/pnas.100422497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hartmann G, Honikel KO, Knusel F, Nuesch J. The specific inhibition of the DNA-directed RNA synthesis by rifamycin. Biochim Biophys Acta. 1967;145:843–4. doi: 10.1016/0005-2787(67)90147-5. [DOI] [PubMed] [Google Scholar]
- 42•.Campbell EA, et al. Structural mechanism for rifampicin inhibition of bacterial rna polymerase. Cell. 2001;104:901–12. doi: 10.1016/s0092-8674(01)00286-0. Describes the intricacies of binding between the rifamycin antibiotic, rifampicin, and a DNA-engaged RNA polymerase, while providing a detailed mechanism for rifamycin action (blockage of the nacent RNA transcript exit channel) based primarially on the results of x-ray crystallography studies. [DOI] [PubMed] [Google Scholar]
- 43.Naryshkina T, Mustaev A, Darst SA, Severinov K. The beta subunit of Escherichia coli RNA polymerase is not required for interaction with initiating nucleotide but is necessary for interaction with rifampicin. J Biol Chem. 2001;276:13308–13. doi: 10.1074/jbc.M011041200. [DOI] [PubMed] [Google Scholar]
- 44.Chamberlin M, Losick R Cold Spring Harbor Laboratory. RNA polymerase. Cold Spring Harbor Laboratory; Cold Spring Harbor, N. Y: 1976. [Google Scholar]
- 45.McClure WR, Cech CL. On the mechanism of rifampicin inhibition of RNA synthesis. J Biol Chem. 1978;253:8949–56. [PubMed] [Google Scholar]
- 46.Artsimovitch I, Chu C, Lynch AS, Landick R. A new class of bacterial RNA polymerase inhibitor affects nucleotide addition. Science. 2003;302:650–4. doi: 10.1126/science.1087526. [DOI] [PubMed] [Google Scholar]
- 47.Sensi P, Margalith P, Timbal MT. Rifomycin, a new antibiotic; preliminary report. Farmaco Sci. 1959;14:146–7. [PubMed] [Google Scholar]
- 48.Sensi P. History of the development of rifampin. Rev Infect Dis. 1983;5 (Suppl 3):S402–6. doi: 10.1093/clinids/5.supplement_3.s402. [DOI] [PubMed] [Google Scholar]
- 49.Wehrli W. Rifampin: mechanisms of action and resistance. Rev Infect Dis. 1983;5 (Suppl 3):S407–11. doi: 10.1093/clinids/5.supplement_3.s407. [DOI] [PubMed] [Google Scholar]
- 50.Burman WJ, Gallicano K, Peloquin C. Comparative pharmacokinetics and pharmacodynamics of the rifamycin antibacterials. Clin Pharmacokinet. 2001;40:327–41. doi: 10.2165/00003088-200140050-00002. [DOI] [PubMed] [Google Scholar]
- 51.Hobby GL, Lenert TF. The action of rifampin alone and in combination with other antituberculous drugs. Am Rev Respir Dis. 1970;102:462–5. doi: 10.1164/arrd.1970.102.3.462. [DOI] [PubMed] [Google Scholar]
- 52•.Kono Y. Oxygen Enhancement of bactericidal activity of rifamycin SV on Escherichia coli and aerobic oxidation of rifamycin SV to rifamycin S catalyzed by manganous ions: the role of superoxide. J Biochem (Tokyo) 1982;91:381–95. doi: 10.1093/oxfordjournals.jbchem.a133698. Reveals that redox cycling of rifamycin drug molecules results in the formation of reactive oxygen species, and that reactive oxygen species generation contributes to the bactericidal activity of the antibiotic. [DOI] [PubMed] [Google Scholar]
- 53.Scrutton MC. Divalent metal ion catalysis of the oxidation of rifamycin SV to rifamycin S. FEBS Lett. 1977;78:216–20. doi: 10.1016/0014-5793(77)80309-8. [DOI] [PubMed] [Google Scholar]
- 54.Bugg TD, Walsh CT. Intracellular steps of bacterial cell wall peptidoglycan biosynthesis: enzymology, antibiotics, and antibiotic resistance. Nat Prod Rep. 1992;9:199–215. doi: 10.1039/np9920900199. [DOI] [PubMed] [Google Scholar]
- 55.Holtje JV. Growth of the stress-bearing and shape-maintaining murein sacculus of Escherichia coli. Microbiol Mol Biol Rev. 1998;62:181–203. doi: 10.1128/mmbr.62.1.181-203.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Park JT, Uehara T. How bacteria consume their own exoskeletons (turnover and recycling of cell wall peptidoglycan) Microbiol Mol Biol Rev. 2008;72:211–27. doi: 10.1128/MMBR.00027-07. table of contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57•.Wise EM, Jr, Park JT. Penicillin: its basic site of action as an inhibitor of a peptide cross-linking reaction in cell wall mucopeptide synthesis. Proc Natl Acad Sci U S A. 1965;54:75–81. doi: 10.1073/pnas.54.1.75. References 56 and 57 describe the results of complementary studies first revealing that inhibition of cell wall biosynthesis by beta-lactam antibiotics is due to catalytic site modification of transpeptidase and carboxypeptidase enzymes (later penicillin binding proteins), which misrecognize the drug molecule as a peptidoglycan substrate mimic. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58•.Tipper DJ, Strominger JL. Mechanism of action of penicillins: a proposal based on their structural similarity to acyl-D-alanyl-D-alanine. Proc Natl Acad Sci U S A. 1965;54:1133–41. doi: 10.1073/pnas.54.4.1133. References 56 and 57 describe the results of complementary studies first revealing that inhibition of cell wall biosynthesis by beta-lactam antibiotics is due to catalytic site modification of transpeptidase and carboxypeptidase enzymes (later penicillin binding proteins), which misrecognize the drug molecule as a peptidoglycan substrate mimic. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Waxman DJ, Yocum RR, Strominger JL. Penicillins and cephalosporins are active site-directed acylating agents: evidence in support of the substrate analogue hypothesis. Philos Trans R Soc Lond B Biol Sci. 1980;289:257–71. doi: 10.1098/rstb.1980.0044. [DOI] [PubMed] [Google Scholar]
- 60.Josephine HR, Kumar I, Pratt RF. The perfect penicillin? Inhibition of a bacterial DD-peptidase by peptidoglycan-mimetic beta-lactams. J Am Chem Soc. 2004;126:8122–3. doi: 10.1021/ja048850s. [DOI] [PubMed] [Google Scholar]
- 61.Kahne D, Leimkuhler C, Lu W, Walsh C. Glycopeptide and lipoglycopeptide antibiotics. Chem Rev. 2005;105:425–48. doi: 10.1021/cr030103a. [DOI] [PubMed] [Google Scholar]
- 62.Cooper MA, Williams DH. Binding of glycopeptide antibiotics to a model of a vancomycin-resistant bacterium. Chem Biol. 1999;6:891–9. doi: 10.1016/s1074-5521(00)80008-3. [DOI] [PubMed] [Google Scholar]
- 63.Ge M, et al. Vancomycin derivatives that inhibit peptidoglycan biosynthesis without binding D-Ala-D-Ala. Science. 1999;284:507–11. doi: 10.1126/science.284.5413.507. [DOI] [PubMed] [Google Scholar]
- 64•.Tomasz A, Albino A, Zanati E. Multiple antibiotic resistance in a bacterium with suppressed autolytic system. Nature. 1970;227:138–40. doi: 10.1038/227138a0. Demonstrates for the first time that beta-lactam-induced cell lysis is regulated by the activity of murein hydrolases. Also reveals that wild-type pneumococci and lysis-defective, murein hydrolase activity-deficient pneumococci are equally sensitive to beta-lactam treatment despite starkly different phenotypic effects. [DOI] [PubMed] [Google Scholar]
- 65•.Heidrich C, Ursinus A, Berger J, Schwarz H, Holtje JV. Effects of multiple deletions of murein hydrolases on viability, septum cleavage, and sensitivity to large toxic molecules in Escherichia coli. J Bacteriol. 2002;184:6093–9. doi: 10.1128/JB.184.22.6093-6099.2002. Reveals that murein hydrolases in E. coli are important for cell separation following cell division and demonstrates that the deletion of multiple murein hydrolase enzymes delays beta-lactam-induced lysis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Uehara T, Dinh T, Bernhardt TG. LytM-domain factors are required for daughter cell separation and rapid ampicillin-induced lysis in Escherichia coli. J Bacteriol. 2009;191:5094–107. doi: 10.1128/JB.00505-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67•.Moreillon P, Markiewicz Z, Nachman S, Tomasz A. Two bactericidal targets for penicillin in pneumococci: autolysis-dependent and autolysis-independent killing mechanisms. Antimicrob Agents Chemother. 1990;34:33–9. doi: 10.1128/aac.34.1.33. Describes the characterization of the cid system in pneumococci, which contributes to killing by beta-lactams independently of murein hydrolase (autolysin) activity. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hoch JA. Two-component and phosphorelay signal transduction. Curr Opin Microbiol. 2000;3:165–70. doi: 10.1016/s1369-5274(00)00070-9. [DOI] [PubMed] [Google Scholar]
- 69.Novak R, Henriques B, Charpentier E, Normark S, Tuomanen E. Emergence of vancomycin tolerance in Streptococcus pneumoniae. Nature. 1999;399:590–3. doi: 10.1038/21202. [DOI] [PubMed] [Google Scholar]
- 70.Novak R, Charpentier E, Braun JS, Tuomanen E. Signal transduction by a death signal peptide: uncovering the mechanism of bacterial killing by penicillin. Mol Cell. 2000;5:49–57. doi: 10.1016/s1097-2765(00)80402-5. [DOI] [PubMed] [Google Scholar]
- 71.Brunskill EW, Bayles KW. Identification and molecular characterization of a putative regulatory locus that affects autolysis in Staphylococcus aureus. J Bacteriol. 1996;178:611–8. doi: 10.1128/jb.178.3.611-618.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Brunskill EW, Bayles KW. Identification of LytSR-regulated genes from Staphylococcus aureus. J Bacteriol. 1996;178:5810–2. doi: 10.1128/jb.178.19.5810-5812.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Groicher KH, Firek BA, Fujimoto DF, Bayles KW. The Staphylococcus aureus lrgAB operon modulates murein hydrolase activity and penicillin tolerance. J Bacteriol. 2000;182:1794–801. doi: 10.1128/jb.182.7.1794-1801.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74•.Rice KC, et al. The Staphylococcus aureus cidAB operon: evaluation of its role in regulation of murein hydrolase activity and penicillin tolerance. J Bacteriol. 2003;185:2635–43. doi: 10.1128/JB.185.8.2635-2643.2003. Suggests that CidAB and LrgAB function as a holin/anti-holin-like system that regulates that activity of murein hydrolases, and subsequently, tolerance to beta-lactam treatment. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Bayles KW. The biological role of death and lysis in biofilm development. Nat Rev Microbiol. 2007;5:721–6. doi: 10.1038/nrmicro1743. [DOI] [PubMed] [Google Scholar]
- 76.Spratt BG. Distinct penicillin binding proteins involved in the division, elongation, and shape of Escherichia coli K12. Proc Natl Acad Sci U S A. 1975;72:2999–3003. doi: 10.1073/pnas.72.8.2999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kitano K, Tomasz A. Triggering of autolytic cell wall degradation in Escherichia coli by beta-lactam antibiotics. Antimicrob Agents Chemother. 1979;16:838–48. doi: 10.1128/aac.16.6.838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lewin CS, Howard BM, Ratcliffe NT, Smith JT. 4-quinolones and the SOS response. J Med Microbiol. 1989;29:139–44. doi: 10.1099/00222615-29-2-139. [DOI] [PubMed] [Google Scholar]
- 79.Bi E, Lutkenhaus J. Cell division inhibitors SulA and MinCD prevent formation of the FtsZ ring. J Bacteriol. 1993;175:1118–25. doi: 10.1128/jb.175.4.1118-1125.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Goehring NW, Beckwith J. Diverse paths to midcell: assembly of the bacterial cell division machinery. Curr Biol. 2005;15:R514–26. doi: 10.1016/j.cub.2005.06.038. [DOI] [PubMed] [Google Scholar]
- 81•.Miller C, et al. SOS response induction by beta-lactams and bacterial defense against antibiotic lethality. Science. 2004;305:1629–31. doi: 10.1126/science.1101630. Describes observations made in E. coli that β-lactam antibiotics can uniquely stimulate expression of the SOS stress response via activation of the DpiBA two-component signal transduction system, and suggests that SOS-mediated arrest of cell division may be a protective reaction to transpeptidase inactivation by these drugs. [DOI] [PubMed] [Google Scholar]
- 82.Varma A, Young KD. FtsZ collaborates with penicillin binding proteins to generate bacterial cell shape in Escherichia coli. J Bacteriol. 2004;186:6768–74. doi: 10.1128/JB.186.20.6768-6774.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Vincent S, Glauner B, Gutmann L. Lytic effect of two fluoroquinolones, ofloxacin and pefloxacin, on Escherichia coli W7 and its consequences on peptidoglycan composition. Antimicrob Agents Chemother. 1991;35:1381–5. doi: 10.1128/aac.35.7.1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Garrett RA. The ribosome: structure, function, antibiotics, and cellular interactions. ASM Press; Washington, DC: 2000. [Google Scholar]
- 85.Nissen P, Hansen J, Ban N, Moore PB, Steitz TA. The structural basis of ribosome activity in peptide bond synthesis. Science. 2000;289:920–30. doi: 10.1126/science.289.5481.920. [DOI] [PubMed] [Google Scholar]
- 86.Katz L, Ashley GW. Translation and protein synthesis: macrolides. Chem Rev. 2005;105:499–528. doi: 10.1021/cr030107f. [DOI] [PubMed] [Google Scholar]
- 87.Mukhtar TA, Wright GD. Streptogramins, oxazolidinones, and other inhibitors of bacterial protein synthesis. Chem Rev. 2005;105:529–42. doi: 10.1021/cr030110z. [DOI] [PubMed] [Google Scholar]
- 88.Patel U, et al. Oxazolidinones mechanism of action: inhibition of the first peptide bond formation. J Biol Chem. 2001;276:37199–205. doi: 10.1074/jbc.M102966200. [DOI] [PubMed] [Google Scholar]
- 89.Vannuffel P, Cocito C. Mechanism of action of streptogramins and macrolides. Drugs. 1996;51 (Suppl 1):20–30. doi: 10.2165/00003495-199600511-00006. [DOI] [PubMed] [Google Scholar]
- 90.Menninger JR, Otto DP. Erythromycin, carbomycin, and spiramycin inhibit protein synthesis by stimulating the dissociation of peptidyl-tRNA from ribosomes. Antimicrob Agents Chemother. 1982;21:811–8. doi: 10.1128/aac.21.5.811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91•.Tenson T, Lovmar M, Ehrenberg M. The mechanism of action of macrolides, lincosamides and streptogramin B reveals the nascent peptide exit path in the ribosome. J Mol Biol. 2003;330:1005–14. doi: 10.1016/s0022-2836(03)00662-4. Reveals that 50S ribosomal subunit binding drugs of the macrolide, lincosamide and streptogramin B classes allow for elongation of distinct amino acid chain lengths during translation, which are determined by the fit between drug molecule and the peptidyltransferase center of the ribosome, before forcing dissociation of the nacent peptidyl-tRNA. [DOI] [PubMed] [Google Scholar]
- 92.Chopra I, Roberts M. Tetracycline antibiotics: mode of action, applications, molecular biology, and epidemiology of bacterial resistance. Microbiol Mol Biol Rev. 2001;65:232–60. doi: 10.1128/MMBR.65.2.232-260.2001. second page, table of contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Davis BD. Mechanism of bactericidal action of aminoglycosides. Microbiol Rev. 1987;51:341–50. doi: 10.1128/mr.51.3.341-350.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Weisblum B, Davies J. Antibiotic inhibitors of the bacterial ribosome. Bacteriol Rev. 1968;32:493–528. [PMC free article] [PubMed] [Google Scholar]
- 95.Hancock RE. Aminoglycoside uptake and mode of action--with special reference to streptomycin and gentamicin. I. Antagonists and mutants. J Antimicrob Chemother. 1981;8:249–76. doi: 10.1093/jac/8.4.249. [DOI] [PubMed] [Google Scholar]
- 96•.Davies J, Gorini L, Davis BD. Misreading of RNA codewords induced by aminoglycoside antibiotics. Mol Pharmacol. 1965;1:93–106. Describes the results of detailed studies which determined the degree of mistranslation and types of mutagenesis induced by various aminoglycoside drugs while the genetic code was first being deciphered. [PubMed] [Google Scholar]
- 97.Karimi R, Ehrenberg M. Dissociation rate of cognate peptidyl-tRNA from the A-site of hyper-accurate and error-prone ribosomes. Eur J Biochem. 1994;226:355–60. doi: 10.1111/j.1432-1033.1994.tb20059.x. [DOI] [PubMed] [Google Scholar]
- 98.Fourmy D, Recht MI, Blanchard SC, Puglisi JD. Structure of the A site of Escherichia coli 16S ribosomal RNA complexed with an aminoglycoside antibiotic. Science. 1996;274:1367–71. doi: 10.1126/science.274.5291.1367. [DOI] [PubMed] [Google Scholar]
- 99.Pape T, Wintermeyer W, Rodnina MV. Conformational switch in the decoding region of 16S rRNA during aminoacyl-tRNA selection on the ribosome. Nat Struct Biol. 2000;7:104–7. doi: 10.1038/72364. [DOI] [PubMed] [Google Scholar]
- 100.Rahal JJ, Jr, Simberkoff MS. Bactericidal and bacteriostatic action of chloramphenicol against memingeal pathogens. Antimicrob Agents Chemother. 1979;16:13–18. doi: 10.1128/aac.16.1.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Goldstein FW, Emirian MF, Coutrot A, Acar JF. Bacteriostatic and bactericidal activity of azithromycin against Haemophilus influenzae. J Antimicrob Chemother. 1990;25 (Suppl A):25–8. doi: 10.1093/jac/25.suppl_a.25. [DOI] [PubMed] [Google Scholar]
- 102.Roberts E, Sethi A, Montoya J, Woese CR, Luthey-Schulten Z. Molecular signatures of ribosomal evolution. Proc Natl Acad Sci U S A. 2008;105:13953–8. doi: 10.1073/pnas.0804861105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Davis BD, Chen LL, Tai PC. Misread protein creates membrane channels: an essential step in the bactericidal action of aminoglycosides. Proc Natl Acad Sci U S A. 1986;83:6164–8. doi: 10.1073/pnas.83.16.6164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Arrow AS, Taber HW. Streptomycin accumulation by Bacillus subtilis requires both a membrane potential and cytochrome aa3. Antimicrob Agents Chemother. 1986;29:141–6. doi: 10.1128/aac.29.1.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105•.Bryan LE, Kwan S. Roles of ribosomal binding, membrane potential, and electron transport in bacterial uptake of streptomycin and gentamicin. Antimicrob Agents Chemother. 1983;23:835–45. doi: 10.1128/aac.23.6.835. Discusses the role of respiration in the uptake of aminoglycosides, as well as the effects of feedback on respiratory activity upon initial drug molecule-target binding. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Hancock R. Uptake of 14C-streptomycin by some microorganisms and its relation to their streptomycin sensitivity. J Gen Microbiol. 1962;28:493–501. doi: 10.1099/00221287-28-3-493. [DOI] [PubMed] [Google Scholar]
- 107.Kogut M, Lightbrown JW, Isaacson P. Streptomycin Action and Anaerobiosis. J Gen Microbiol. 1965;39:155–64. doi: 10.1099/00221287-39-2-155. [DOI] [PubMed] [Google Scholar]
- 108.Bryan LE, Kowand SK, Van Den Elzen HM. Mechanism of aminoglycoside antibiotic resistance in anaerobic bacteria: Clostridium perfringens and Bacteroides fragilis. Antimicrob Agents Chemother. 1979;15:7–13. doi: 10.1128/aac.15.1.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Anand N, Davis BD. Damage by streptomycin to the cell membrane of Escherichia coli. Nature. 1960;185:22–3. doi: 10.1038/185022a0. [DOI] [PubMed] [Google Scholar]
- 110.Anand N, Davis BD, Armitage AK. Uptake of streptomycin by Escherichia coli. Nature. 1960;185:23–4. doi: 10.1038/185023a0. [DOI] [PubMed] [Google Scholar]
- 111.Ruiz N, Silhavy TJ. Sensing external stress: watchdogs of the Escherichia coli cell envelope. Curr Opin Microbiol. 2005;8:122–6. doi: 10.1016/j.mib.2005.02.013. [DOI] [PubMed] [Google Scholar]
- 112.Liu X, De Wulf P. Probing the ArcA-P modulon of Escherichia coli by whole genome transcriptional analysis and sequence recognition profiling. J Biol Chem. 2004;279:12588–97. doi: 10.1074/jbc.M313454200. [DOI] [PubMed] [Google Scholar]
- 113.Malpica R, Franco B, Rodriguez C, Kwon O, Georgellis D. Identification of a quinone-sensitive redox switch in the ArcB sensor kinase. Proc Natl Acad Sci U S A. 2004;101:13318–23. doi: 10.1073/pnas.0403064101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114•.Brazas MD, Hancock RE. Using microarray gene signatures to elucidate mechanisms of antibiotic action and resistance. Drug Discov Today. 2005;10:1245–52. doi: 10.1016/S1359-6446(05)03566-X. Discusses the utility of studying gene expression signatures (or patterns in gene expression), derived from microarray-based studies of antibiotic-treated bacteria, in efforts to uncover novel drug targets and off-target effects that contribute to drug-induced cell death. [DOI] [PubMed] [Google Scholar]
- 115.Tamae C, et al. Determination of antibiotic hypersensitivity among 4,000 single-gene-knockout mutants of Escherichia coli. J Bacteriol. 2008;190:5981–8. doi: 10.1128/JB.01982-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Breidenstein EB, Khaira BK, Wiegand I, Overhage J, Hancock RE. Complex ciprofloxacin resistome revealed by screening a Pseudomonas aeruginosa mutant library for altered susceptibility. Antimicrob Agents Chemother. 2008;52:4486–91. doi: 10.1128/AAC.00222-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Dwyer DJ, Kohanski MA, Collins JJ. Networking opportunities for bacteria. Cell. 2008;135:1153–6. doi: 10.1016/j.cell.2008.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Faith JJ, et al. Large-scale mapping and validation of Escherichia coli transcriptional regulation from a compendium of expression profiles. PLoS Biol. 2007;5:e8. doi: 10.1371/journal.pbio.0050008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Gardner TS, di Bernardo D, Lorenz D, Collins JJ. Inferring genetic networks and identifying compound mode of action via expression profiling. Science. 2003;301:102–5. doi: 10.1126/science.1081900. [DOI] [PubMed] [Google Scholar]
- 120.Bonneau R, et al. A predictive model for transcriptional control of physiology in a free living cell. Cell. 2007;131:1354–65. doi: 10.1016/j.cell.2007.10.053. [DOI] [PubMed] [Google Scholar]
- 121.Ronen M, Rosenberg R, Shraiman BI, Alon U. Assigning numbers to the arrows: parameterizing a gene regulation network by using accurate expression kinetics. Proc Natl Acad Sci U S A. 2002;99:10555–60. doi: 10.1073/pnas.152046799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Feist AM, Herrgard MJ, Thiele I, Reed JL, Palsson BO. Reconstruction of biochemical networks in microorganisms. Nat Rev Microbiol. 2009;7:129–43. doi: 10.1038/nrmicro1949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Imlay JA. Pathways of oxidative damage. Annu Rev Microbiol. 2003;57:395–418. doi: 10.1146/annurev.micro.57.030502.090938. [DOI] [PubMed] [Google Scholar]
- 124.Schurek KN, et al. Novel genetic determinants of low-level aminoglycoside resistance in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 2008;52:4213–9. doi: 10.1128/AAC.00507-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Wigle TJ, et al. Inhibitors of RecA activity discovered by high-throughput screening: cell-permeable small molecules attenuate the SOS response in Escherichia coli. J Biomol Screen. 2009;14:1092–101. doi: 10.1177/1087057109342126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Imlay JA. How oxygen damages microbes: oxygen tolerance and obligate anaerobiosis. Adv Microb Physiol. 2002;46:111–53. doi: 10.1016/s0065-2911(02)46003-1. [DOI] [PubMed] [Google Scholar]
- 127.Davies BW, et al. Hydroxyurea induces hydroxyl radical-mediated cell death in Escherichia coli. Mol Cell. 2009;36:845–60. doi: 10.1016/j.molcel.2009.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Dwyer DJ, Kohanski MA, Collins JJ. Role of reactive oxygen species in antibiotic action and resistance. Curr Opin Microbiol. 2009;12:482–9. doi: 10.1016/j.mib.2009.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Kohanski MA, DePristo MA, Collins JJ. Sub-lethal antibiotic treatment leads to multidrug resistance via radical-induced mutagenesis. Molecular Cell. 2010;XX:XX–XX. doi: 10.1016/j.molcel.2010.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Gusarov I, Shatalin K, Starodubtseva M, Nudler E. Endogenous nitric oxide protects bacteria against a wide spectrum of antibiotics. Science. 2009;325:1380–4. doi: 10.1126/science.1175439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Vazquez-Torres A, et al. Salmonella pathogenicity island 2-dependent evasion of the phagocyte NADPH oxidase. Science. 2000;287:1655–8. doi: 10.1126/science.287.5458.1655. [DOI] [PubMed] [Google Scholar]
- 132.Eggert US, et al. Genetic basis for activity differences between vancomycin and glycolipid derivatives of vancomycin. Science. 2001;294:361–4. doi: 10.1126/science.1063611. [DOI] [PubMed] [Google Scholar]
- 133.Muthaiyan A, Silverman JA, Jayaswal RK, Wilkinson BJ. Transcriptional profiling reveals that daptomycin induces the Staphylococcus aureus cell wall stress stimulon and genes responsive to membrane depolarization. Antimicrob Agents Chemother. 2008;52:980–90. doi: 10.1128/AAC.01121-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Hancock RE, Rozek A. Role of membranes in the activities of antimicrobial cationic peptides. FEMS Microbiol Lett. 2002;206:143–9. doi: 10.1111/j.1574-6968.2002.tb11000.x. [DOI] [PubMed] [Google Scholar]
- 135.Hancock RE, Sahl HG. Antimicrobial and host-defense peptides as new anti-infective therapeutic strategies. Nat Biotechnol. 2006;24:1551–7. doi: 10.1038/nbt1267. [DOI] [PubMed] [Google Scholar]
- 136.Keith CT, Borisy AA, Stockwell BR. Multicomponent therapeutics for networked systems. Nat Rev Drug Discov. 2005;4:71–8. doi: 10.1038/nrd1609. [DOI] [PubMed] [Google Scholar]
- 137.Yeh PJ, Hegreness MJ, Aiden AP, Kishony R. Drug interactions and the evolution of antibiotic resistance. Nat Rev Microbiol. 2009;7:460–6. doi: 10.1038/nrmicro2133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138•.Yeh P, Tschumi AI, Kishony R. Functional classification of drugs by properties of their pairwise interactions. Nat Genet. 2006;38:489–94. doi: 10.1038/ng1755. Demonstrates a quantitative, network-based approach for studying drug-drug interactions that also allows for the elucidation of the functional mechanisms underlying drug mode-of-action and affected cellular targets. [DOI] [PubMed] [Google Scholar]
- 139.Bollenbach T, Quan S, Chait R, Kishony R. Nonoptimal microbial response to antibiotics underlies suppressive drug interactions. Cell. 2009;139:707–18. doi: 10.1016/j.cell.2009.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Plotz PH, Davis BD. Synergism between streptomycin and penicillin: a proposed mechanism. Science. 1962;135:1067–8. doi: 10.1126/science.135.3508.1067. [DOI] [PubMed] [Google Scholar]
- 141.Lu TK, Collins JJ. Dispersing biofilms with engineered enzymatic bacteriophage. Proc Natl Acad Sci U S A. 2007;104:11197–202. doi: 10.1073/pnas.0704624104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Lu TK, Collins JJ. Engineered bacteriophage targeting gene networks as adjuvants for antibiotic therapy. Proc Natl Acad Sci U S A. 2009;106:4629–34. doi: 10.1073/pnas.0800442106. [DOI] [PMC free article] [PubMed] [Google Scholar]