Abstract
Lung cancer is the leading cause of cancer deaths in the U.S., surpassing breast cancer as the primary cause of cancer-related mortality in women. The goal of the present study was to identify early molecular changes in the lung induced by exposure to tobacco smoke and thus identify potential targets for chemoprevention. Female A/J mice were exposed to either tobacco smoke or HEPA-filtered air via a whole-body exposure chamber (6 h/day; 5 days/wk for 3, 8 and 20 wk). Gene expression profiles of lung tissue from control and smoke-exposed animals were established using a 15 K cDNA microarray. Cytochrome P450 1b1 (Cyp1b1), a Phase I enzyme involved in both the metabolism of xenobiotics and the 4-hydroxylation of 17β-estradiol, was modulated to the greatest extent following smoke exposure. A panel of 10 genes was found to be differentially expressed in control and smoke-exposed lung tissue at 3, 8 and 20 wk (P < 0.001). The interaction network of these differentially expressed genes revealed new pathways modulated by short-term smoke exposure including estrogen metabolism. In addition, 17β-estradiol was detected within murine lung tissue by gas chromatography coupled mass spectrometry and immunohistochemistry. Identification of the early molecular events that contribute to lung tumor formation is anticipated to lead to the development of promising targeted chemopreventive therapies. In conclusion, the presence of 17β-estradiol within lung tissue when combined with the modulation of Cyp1b1 and other estrogen metabolism genes by tobacco smoke provides novel insight into a possible role for estrogens in lung cancer.
Keywords: Tobacco smoke, mouse lung, gene expression, Cyp1b1, estrogen
Lung cancer is the leading cause of cancer death in the U.S. and has surpassed breast cancer as the primary cause of cancer-related mortality in women (1). Exposure to cigarette smoke is estimated to account for approximately 90% of all lung cancers (2). Epidemiologic and clinical data suggest a gender difference in the biology of lung cancer (3, 4). Women appear to have an increased susceptibility to tobacco carcinogens but have a better prognosis after lung cancer diagnosis as compared to men (3). It has been suggested that estrogens may affect the susceptibility of women to lung cancer (2, 5-7).
The individual variation in efficiency of cellular processes such as detoxification, metabolic activation, adduct formation and DNA repair influences individual susceptibility to the tumorigenic effects associated with tobacco smoke exposure. Detoxification enzymes play a critical role in the metabolism of both tobacco-related carcinogens and endogenous compounds (including hormones) within the human lung (8). Activation of the aryl hydrocarbon receptor (AHR) signaling pathway by components of tobacco smoke such as polycyclic aromatic hydrocarbons (PAHs) leads to transcriptional upregulation of a number of genes including members of the cytochrome P450 (CYP) family, in particular CYP1B1 and CYP1A1. These Phase I enzymes activate procarcinogens like benzo(a)pyrene (BaP) to reactive electrophilic intermediates, which are in general detoxified to inactive water-soluble conjugates for excretion by Phase II enzymes (i.e., glutathione S-transferase (GST), UDP-glucuronosyl-transferase (UGT), sulfotransferase (SULT) and catechol-O-methyltransferase (COMT)). Up- or downregulation of these enzymes, in turn, can influence the fate of reactive intermediates in the body, leading to the accumulation or clearance of metabolites; thus influencing cancer incidence.
It is well established that estrogen can stimulate cell signaling and proliferation in the breast via estrogen receptor (ER)-dependent and -independent pathways (9). The role of estrogen as a procarcinogen is also based on the ability of its metabolites (catechol estrogens) to induce genetic damage. The formation of catechol estrogens is catalyzed by CYP1B1, CYP1A1, CYP1A2 and CYP3A4 in several tissues. The most carcinogenic catechol estrogen is 4-hydroxyestradiol (4-E2), an estrogen agonist that binds to the ER with greater affinity and for longer periods of time than the parent compound 17β-estradiol (E2) (10). 4-E2 is produced primarily by CYP1B1 and is quickly oxidized to highly reactive quinones that bind to DNA, forming depurinating DNA adducts (11) that induce genetic mutations.
Although smoking cessation decreases the risk of lung cancer, ex-smokers remain at a significantly increased risk for cancer for decades. Development of an efficacious chemopreventive regimen for lung cancer has been hindered by our inability to identify early molecular targets for intervention as well as those individuals who would benefit most from treatment. The majority of the molecular targets that have been identified to date represent late events in tumorigenesis, as indicated by their presence in established tumors. These include biomarkers of tumor cell proliferation (i.e., EGFR, TP53, KRAS, RB, BCL2) and angiogenesis-stimulation factors (i.e., VEGF, FGF, MMP), among others (12). While these late events aid in selecting treatment options and predicting prognosis, early events (in particular, those occurring during the preneoplastic phase) can be used as targets for chemopreventive intervention, thus blocking tumor formation. Limited attention has been given to the identification of early biomarkers of lung cancer risk.
The A/J mouse and its genetically related A/HeJ strain have been shown to be the strains that are most responsive to cigarette smoke exposure as compared to AKR, BALB/C, C3H/HeJ, C57BL/6, CAST/Ei, DBA, SWR, and 129/Svi mice (13). A/J mice develop lung adenomas/adenocarcinomas spontaneously and, following exposure to tobacco smoke, readily develop additional lung tumors that are similar to human lung adenocarcinomas with respect to pathological features, genetic alterations and aberrant signaling pathways (14). By allowing the animals to recover in fresh air for 16 wk following 20 wk of smoke exposure, Witschi and colleagues (15) were successful in enhancing lung tumor development in smoke-exposed mice.
The goal of the present study was to identify early changes in gene expression that are induced within the nonneoplastic lung tissue of female A/J mice following exposure to tobacco smoke. Such early alterations in gene expression may yield insight into tumor initiation and promotion, and represent both early biomarkers of lung cancer development and molecular targets for chemopreventive intervention. In this study, we compared the gene expression profile of lung tissue from mice exposed to tobacco smoke for 3-20 wk with that of lungs from control animals maintained in ambient air for the same period of time. Our enhanced understanding of the early molecular events that contribute to lung tumor development is anticipated to aid in the identification of promising chemopreventive targets that can be examined readily for their efficacy in the A/J model of current and former smoke exposure.
Materials and Methods
Tobacco smoke exposure
Female A/J mice, 8-9 wk of age, were purchased from the Jackson Laboratory (Bar Harbor, ME) and maintained with free access to food (AIN-93G, Harlan Teklad Global Diets, Indianapolis, IN) and water. Body weights were recorded weekly to monitor growth. All procedures were approved by the Institutional Animal Care and Use Committee of the University of Kentucky, Lexington, KY.
Mice were randomized into 6 treatment groups (6 per group) and exposed to either tobacco smoke (S) or HEPA-filtered ambient air (control) (C). Exposure of mice to sidestream smoke was carried out in whole-body chambers for 6 h per day, 5 days per wk for 3, 8 and 20 wk. Sidestream smoke was generated from the University of Kentucky reference cigarette (2R4F), and the suspended smoke particulate concentration in the chamber atmosphere was maintained at approximately 40-45 mg/m3.
Exposure of mice to smoke was ascertained by measuring levels of urinary cotinine, ethoxyresorufin deethylase (EROD) activity in lung microsomes and cytochrome P450 1a1 (Cyp1a1) protein. Significant increases of these markers were observed in smoke-exposed groups over controls at all time points. Cotinine was measured routinely in urine samples (ELISA Kit, OraSure Technologies, Bethlehem, PA) collected from 3-5 mice/group over 24 h. Cotinine values ranging from 4-10 μg cotinine/mg creatinine were observed in urine from smoke-exposed mice as compared to picogram levels in the unexposed control groups. Lobes of lungs from 3-4 animals per treatment group were pooled, and lung microsomes were prepared by differential centrifugation (9,000 ×g and then 100,000 ×g for 60 min). The protein content of the resulting fraction was determined using a commercial kit from Bio-Rad (Hercules, CA). The EROD activity of lung microsomes was determined using a modified fluorimetric resorufin assay (16). Aliquots of the microsomal protein were separated on 12% SDS-polyacrylamide gels and subjected to Western blot analysis. Blots were probed with a polyclonal rabbit anti-rat Cyp1a1 antibody (diluted 1:800) from XenoTech LLC (Lenexa, KS) and secondary anti-rabbit antibody conjugated with alkaline phosphatase. Typical EROD activity levels (mean ± standard error of the mean) were 1.21 ± 0.3 and 9.74 ± 1.4 pmoles/min/mg protein for control and smoke-exposed groups, respectively. Immunoblots of lung microsomes probed with anti-Cyp1a1 antibody also showed significantly higher levels of protein in smoke-exposed mice.
At the time of sacrifice, lung tissue was excised from each animal, immediately placed in TRIzol® (Invitrogen Corp., Carlsbad, CA) and stored at −80°C for subsequent RNA isolation and analysis. The remaining lung tissue was snap-frozen in liquid nitrogen for Cyp1a1 analyses.
RNA extraction and probe preparation
Frozen tissues were homogenized in TRIzol® using a Polytron® System PT 1200C (Kinematica AG, Switzerland), and total cellular RNA was extracted as recommended by the manufacturer. RNA concentration was determined by absorbance at 260 nm, and quality was assessed by monitoring the integrity of the 28S and 18S ribosomal RNA by agarose gel electrophoresis.
Equal amounts of RNA from 3 animals were pooled, yielding 2 control and 2 smoke-exposed RNA pools per time point (3 and 8 wk). In order to circumvent the potential contamination of normal lung tissue with neoplastic cells, samples collected at 20 wk were evaluated individually, totaling 6 control and 6 smoke-exposed samples.
A total of 20 pooled (3 and 8 wk) and individual (20 wk) RNA samples (1 μg each) were subjected to one round of T7 based linear RNA amplification using the RiboAmp Kit (Molecular Devices, Sunnyvale, CA). Universal mouse total RNA (Clontech Laboratories, Inc., Palo Alto, CA) was also subjected to one round of amplification and served as a common reference for all hybridizations. cDNA probes were synthesized in duplicate by a standard reverse transcription reaction using 2 μg of each amplified RNA (aRNA) and labeled by indirect (amino-allyl) incorporation of Cy3 or Cy5 (CyDye™ Post-Labeling Reactive Dyes, Amersham Biosciences Corp, Piscataway, NJ) (dye-flip replicates). The concentration of the labeled cDNA probe was determined using a ND-1000 Spectrophotometer (NanoDrop Technologies, Inc., Montchanin, DE).
cDNA microarray hybridization
The expression profile of samples was established using a mouse microarray containing 15,552 (15K) cDNA clones obtained from the Institute of Aging, NIH (sequence information is available at http://lgsun.grc.nia.nih.gov/cDNA/15k.html) and printed at Fox Chase Cancer Center. Probe hybridizations were performed following standard procedures. Following hybridization, the slides were scanned with a GMS 428 Scanner (Affymetrix, Santa Clara, CA) at full laser intensity and variable photomultiplier tube voltage settings, capturing the full dynamic range for each slide in each respective channel. Image segmentation and spot quantification were performed with the ImaGene software, Version 5.6.1 (BioDiscovery, Marina del Rey, CA) using the software's original default settings. The mean intensities of signal and local background were extracted for each spot and subjected to analysis.
Mathematical analyses
Microarray data were processed and analyzed using R (www.r-project.org/) and the Bioconductor (17) platform. Only spots with GenBank accession entries (n = 15,245) were considered for analysis. Background correction was carried out using the normexp method (18) implemented in the Bioconductor package limma (Linear Models for Microarray Data), with an offset of 50. This method has been found to be preferable to local background subtraction in most cases. LOWESS (Locally Weighted Regression and Smoothing Scatter) normalization was used to correct for intensity-dependent dye bias. Dye-swap replicates were considered as replicates for statistical comparisons. In order to identify genes that were differentially expressed between smoke-exposed and reference samples, an empirical Bayes moderated t-test, as implemented in limma (19), was used. Due to differences in the sample design, the differential expression analysis was carried out separately for samples at 3 and 8 wk (pooled samples) and 20 wk (individual samples). Lists of differentially expressed genes for downstream analyses were selected using a P value threshold of 0.001.
Ingenuity Pathways Analysis (IPA version 6.3) (http://www.ingenuity.com/) was used to search for underlying biological pathways and molecular networks. IPA provides a rich functional annotation of genes and proteins and protein-protein interactions as well as the role of genes in various diseases. The genes differentially expressed at all time points (3, 8 and 20 wk) were uploaded into IPA along with the corresponding fold change values. These genes are searched in the IPA's functional annotation database called Ingenuity Pathways Knowledge Database (IPKB). Depending on the input gene list, Ingenuity software models networks and pathways through a statistical computation using functional relationships such as interaction, activation, localization, between proteins, genes, complexes, cells, tissues, drugs and diseases. Given a list of genes and their expression values or fold changes, IPA computes a score (p-value) for network eligible genes. A higher score implies a significant composition of genes in a network.
Gas chromatography coupled mass spectrometry
Female A/J mice (n = 8) at 8 wk of age were purchased from the Jackson Laboratory. At the time of sacrifice, the lung was perfused with 30 ml of saline, excised, and 4 lobes were stored at −80°C for subsequent analysis by gas chromatography/mass spectrometry (GC/MS). Frozen lung tissues were homogenized in 30 mM potassium phosphate buffer (pH 6.0) containing 0.5 mM ascorbic acid. After adding methanol (60% v/v), the homogenate was extracted twice with 1 volume of hexane. The aqueous phase was filtered using a 0.7 micron glass microfiber filter, extracted with 2 volumes of ethyl acetate, and evaporated to dryness. The samples were derivatized in acetonitrile using N,O-bis-(trimethylsilyl)tri-fluoroacetamide containing 1% trimethylchlorosilane. Deuterium-labeled E2 (d5-E2) (C/D/N Isotopes, Pointe-Claire, Quebec, Canada) was used as internal standard and was added prior to splitless injection into a HP6890 GC/MS instrument with capillary column (20m × 0.18mm × 0.18μm, DB-5ms (Agilent JW Scientific Columns, Agilent Technologies, Palo Alto, CA). Selective Ion Monitoring (SIM; m/z 342, 416 and 421 for E1, E2 and d5-E2; respectively) and retention times relative to d5-E2 were used to identify each compound.
Immunohistochemistry
Perfused lungs from three non-ovariectomized female A/J mice were fixed in 10% formalin for 24 h and subsequently embedded in paraffin for immunohistochemical analysis. Sections (4 μm) were dewaxed through incubations in xylene, followed by a graded alcohol series, ending in distilled water. Steam heat-induced epitope recovery (SHIER) was used prior to incubation with the primary antibody. Rabbit polyclonal antibodies for E2 (AR038-5R, Biogenex, San Ramon, CA), ERβ (51-7700, Zymed Laboratories, South San Francisco, CA), and a mouse monoclonal antibody for ERα (clone ER88, Biogenex) were used. All sections were developed using standard immunohistochemical protocols.
Quantitative real-time PCR
Primers specific for each murine gene of interest were purchased from Applied Biosystems, Inc. (Foster City, CA) as follows: Cyp1b1 (assay ID: Mm00487229_m1), Cry1 (assay ID: Mm00514392_m1), Cbr3 (carbonyl reductase 3; assay ID: Mm00557339_m1), Ces3 (carboxylesterase 3; assay ID: Mm00474816_m1), Col3a1 (collagen, type III, alpha 1; assay ID: Mm00802331_m1), Hdc (histidine decarboxylase; assay ID: Mm00456104_m1), Tef (thyrotrophic embryonic factor; assay ID: Mm00457513_m1), Ugt1a6a (UDP-Glycosyltransferase 1 family, polypeptide A6; assay ID: Mm01967851_s1) and Hprt1 (hypoxanthine guanine phosphoribosyl transferase; assay ID: Mm00446968_m1). Total RNA (1 μg) was converted to cDNA using the High Capacity cDNA Archive Kit (Applied Biosystems, Inc.). Quantitative real-time PCR reactions were performed in quadruplicate in an Applied Biosystems 7900HT Fast Real-Time PCR System using universal conditions. Data for each test gene and the housekeeping gene (Hprt1) were obtained in the form of threshold-cycle number (Ct) for each time point (3 wk, 8 wk and 20 wk) and treatment condition (control, smoke-treated). The Ct values for each gene (at each time point) were normalized to the housekeeping gene, and ΔCt values for samples from smoke-treated and control groups were compared using the Mann-Whitney test. The step-up method of Benjamini & Hochberg (20) was employed to account for multiple hypotheses testing, and the False Discovery Rate (FDR) was computed for each gene. An FDR cut-off of 0.10 was used to declare statistical significance. The fold-change in the transcript levels of samples from smoke-treated and control groups was computed at each time point using the comparative Ct method (ΔΔCt) (Applied Biosystems Reference Manual, User Bulletin #2).
Western blot analysis
Fifty micrograms of pulmonary microsomal protein isolated from human smokers and nonsmokers (XenoTech LLC) were separated by 10% sodium dodecyl sulfate polyacrylamide gel electrophoresis (Bio-Rad) and electro-blotted to a polyvinylidene fluoride membrane. Membranes were blocked for 1 h at room temperature in Tris-buffered saline with Tween-20 (TBST) (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 0.1% Tween-20) containing 5% nonfat milk and incubated overnight at 4°C with primary antibodies. Primary antibodies against CYP1B1 and HPRT were purchased from Imgenex Corp. (San Diego, CA) (Catalog No: IMG-5988A) and Abcam, Inc. (Cambridge, MA) (Catalog No: ab10479), respectively. After washing 3 times with TBST, the membranes were incubated with horseradish peroxidase (HRP)-conjugated goat anti-rabbit IgG secondary antibody (Bio-Rad) for 1 h at room temperature, rinsed with TBST and visualized using ECL Western Blotting Detection Reagents (GE Healthcare, Piscataway, NJ).
Results
Genes modulated by tobacco smoke exposure
Using a 15K mouse cDNA array, the global gene expression profile of murine lung tissue from female A/J mice exposed to tobacco smoke was compared to that of age-matched control mice maintained in HEPA-filtered ambient air. After normalization, a strong correlation (r > 0.8) was observed between all dye-swap replicates as well as between 2 pools of samples (n = 3 mice per pool) from the same treatment group (data not shown).
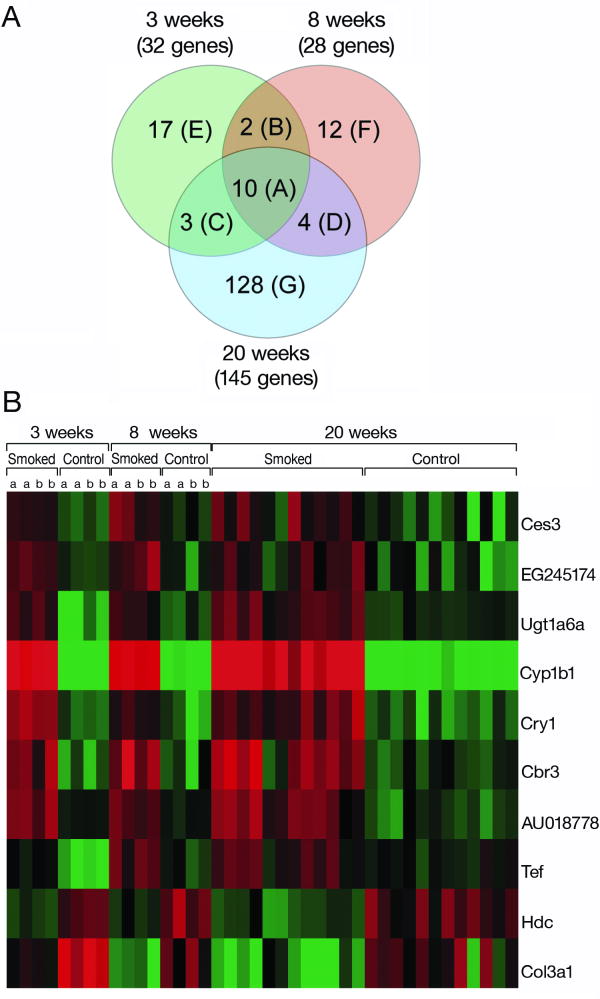
In order to identify early molecular changes induced by tobacco smoke, gene expression profiles from control and smoke-exposed lung tissues were determined following 3, 8 and 20 wk of exposure. The expression of 32, 28, and 145 genes was modulated significantly by smoke following 3, 8 and 20 wk, respectively (P = 0.001) (Fig. 1A). Ten genes were identified as differentially expressed at all time points (Group A, Table 1). The heat map represents fold-change in the expression of these genes following 3, 8 and 20 wk of exposure (Fig. 1B). Hierarchical clustering of the median corrected expression values of this subset of genes showed a precise separation of control and smoke-exposed samples (data not shown). Interestingly, the magnitude of the change in expression was similar for all smoke-exposed groups.
Fig. 1. Genes differentially expressed following 3, 8 and 20 wk of smoke exposure.
A. Venn diagram showing the distribution of differentially expressed genes (P < 0.001) between control lungs and lungs exposed to smoke for 3, 8 and 20 wk. The 10 genes depicted in Group A are modulated to all 3 time points. Groups B-D correspond to genes in common between 3 and 8 wk (B), 3 and 20 wk (C), and 8 and 20 wk (D), whereas Groups E-G represent genes identified only at 3 wk (E), 8 wk (F), or 20 wk (G). The genes represented in each group are listed in Table 1. B. A heat map representing the median normalized expression values for the 10 genes altered at all 3 time points. Data for technical (same letter, i.e. aa) and biological (different letters, i.e. ab) replicates are included.
TABLE 1.
Genes differentially expressed following 3, 8 and 20 weeks of smoke exposurea
| Group A | Group G | ||||
| Gene Symbol | 3 weeks | 8 weeks | 20 weeks | Gene Symbol | 20 weeks |
| Cyp1b1 | 11.38 | 8.19 | 9.81 | Scel | 2.06 |
| Cry1 | 2.66 | 2.04 | 2.18 | Igh-6 | 2.01 |
| Cbr3 | 2.53 | 2.58 | 2.02 | Acsl1 | 1.90 |
| Ugt1a6a | 2.52 | 1.67 | 1.73 | Adprhl2 | 1.85 |
| AU018778 | 1.88 | 1.63 | 2.02 | Nr1d2 | 1.83 |
| EG245174 | 1.85 | 1.90 | 1.58 | Mylpf | 1.81 |
| Ces3 | 1.80 | 1.74 | 1.64 | Cxcl1 | 1.80 |
| Tef | 1.79 | 1.79 | 1.38 | Gclc | 1.78 |
| Hdc | 0.52 | 0.52 | 0.55 | Gpx2 | 1.72 |
| Col3a1 | 0.42 | 0.41 | 0.49 | LOC100039206 | 1.66 |
| 4922501L14Rik | 1.61 | ||||
| Group B | Slc40a1 | 1.58 | |||
| Gene Symbol | 3 weeks | 8 weeks | 20 weeks | Fbln2 | 1.53 |
| Ier3 | 2.00 | 1.86 | - | Gmnn | 1.50 |
| Ttc21a | 0.54 | 0.50 | - | Fabp3 | 1.48 |
| Bclaf1 | 1.48 | ||||
| Group C | Tsix | 1.46 | |||
| Gene Symbol | 3 weeks | 8 weeks | 20 weeks | Cldn12 | 1.45 |
| Akr1c14 | 2.60 | - | 3.02 | Tex12 | 1.44 |
| Pdia6 | 0.64 | - | 0.60 | Cox7a2 | 1.42 |
| Igfbp3 | 0.47 | - | 0.54 | Slc39a4 | 1.41 |
| Bmp1 | 1.41 | ||||
| Group D | Pigc | 1.41 | |||
| Gene Symbol | 3 weeks | 8 weeks | 20 weeks | Ak3l1 | 1.41 |
| Fkbp5 | - | 2.36 | 2.35 | Gas1 | 1.40 |
| Gsta4 | - | 2.02 | 1.81 | 1700020C11Rik | 1.40 |
| Hsp90aa1 | - | 0.58 | 0.68 | Elof1 | 1.40 |
| Ncald | - | 0.54 | 0.71 | LOC100043812 | 1.39 |
| Ndufc1 | 1.39 | ||||
| Group E | Group F | Gsto1 | 1.38 | ||
| Gene Symbol | 3 weeks | Symbol | 8 weeks | Cabc1 | 1.38 |
| Dnajb7 | 2.32 | Gc | 2.53 | Prl3d1 | 1.38 |
| Wipf3 | 1.98 | Serpina1e | 2.28 | Hnrnpu | 1.37 |
| Ppap2b | 1.84 | Tdrd5 | 1.78 | Fancm | 1.37 |
| Gstm1 | 1.64 | Ephx1 | 1.76 | Zfp11 | 1.37 |
| Lonrf3 | 1.64 | Tdrd7 | 1.57 | Apbb2 | 1.35 |
| Tspan15 | 0.63 | Apol6 | 1.56 | Snx14 | 1.34 |
| Igf1 | 0.62 | Slco4c1 | 0.61 | Akr1c19 | 1.34 |
| Col1a2 | 0.60 | Dach1 | 0.53 | Tmem57 | 1.33 |
| Cd34 | 0.58 | Mat2a | 0.52 | Esd | 1.32 |
| Arhgap28 | 0.58 | Axud1 | 0.52 | Josd3 | 1.32 |
| Plekha6 | 0.58 | Errfi1 | 0.50 | Sorbs1 | 1.31 |
| Fbp2 | 0.56 | Pitpnb | 0.38 | Appl2 | 1.31 |
| Npr3 | 0.53 | D19Ertd652e | 1.30 | ||
| Igsf10 | 0.52 | Xdh | 1.30 | ||
| D0H4S114 | 0.47 | Vdac1 | 1.30 | ||
| Slc38a5 | 0.45 | Lias | 1.29 | ||
| Npnt | 0.40 | RP23-292J1.1 | 1.27 | ||
| Bcnp1 | 1.27 | ||||
| 1600020E01Rik | 1.27 | ||||
| Tnk2 | 1.27 | ||||
| Trhde | 1.26 | ||||
| Gtf2a2 | 1.25 | ||||
| Trub2 | 1.25 | ||||
| Bach1 | 1.25 | ||||
| Nfatc4 | 1.24 | ||||
| Zfp612 | 1.24 | ||||
| Krtap16-10 | 1.24 | ||||
| Khdc1a | 1.23 | ||||
| Ncdn | 1.22 | ||||
| Ptp4a1 | 1.21 | ||||
| Pygb | 0.84 | ||||
| Cyhr1 | 0.84 | ||||
| Ptpn6 | 0.83 | ||||
| Maged2 | 0.83 | ||||
| Col4a1 | 0.83 | ||||
| Abca3 | 0.82 | ||||
| Slc15a4 | 0.82 | ||||
| Ankrd17 | 0.81 | ||||
| Itgb7 | 0.81 | ||||
| Ubr5 | 0.80 | ||||
| Wdr79 | 0.79 | ||||
| Srebf1 | 0.79 | ||||
| Rhbdd3 | 0.79 | ||||
| Stmn1 | 0.79 | ||||
| Ubxd2 | 0.79 | ||||
| Ptms | 0.79 | ||||
| Pdia5 | 0.78 | ||||
| Azin1 | 0.78 | ||||
| Zdhhc18 | 0.78 | ||||
| Gm2a | 0.76 | ||||
| Bst2 | 0.75 | ||||
| Rac2 | 0.75 | ||||
| Selplg | 0.75 | ||||
| Diablo | 0.74 | ||||
| Dhrs3 | 0.74 | ||||
| Tia1 | 0.73 | ||||
| Mpv17 | 0.73 | ||||
| Hba-a1 | 0.73 | ||||
| Banp | 0.73 | ||||
| Phtf1 | 0.73 | ||||
| Gcap14 | 0.72 | ||||
| Cybb | 0.72 | ||||
| Stip1 | 0.72 | ||||
| Laptm5 | 0.72 | ||||
| Reps1 | 0.72 | ||||
| Lmbr1 | 0.71 | ||||
| Myo1e | 0.71 | ||||
| Arf1 | 0.71 | ||||
| Bbx | 0.71 | ||||
| Cbx5 | 0.71 | ||||
| Tbc1d15 | 0.71 | ||||
| Sdc4 | 0.70 | ||||
| Comtd1 | 0.70 | ||||
| Xbp1 | 0.70 | ||||
| Lpcat1 | 0.70 | ||||
| Igtp | 0.69 | ||||
| Ywhaq | 0.69 | ||||
| 9030425E11Rik | 0.68 | ||||
| Rnf113a1 | 0.68 | ||||
| Hsp90ab1 | 0.67 | ||||
| Tcfcp2l1 | 0.67 | ||||
| Ipo5 | 0.66 | ||||
| Ahcyl2 | 0.65 | ||||
| Ndst1 | 0.65 | ||||
| LOC100043546 | 0.65 | ||||
| Aspm | 0.64 | ||||
| H2-K1 | 0.64 | ||||
| Hbb-b1 | 0.64 | ||||
| Thra | 0.63 | ||||
| 2810025M15Rik | 0.63 | ||||
| Leo1 | 0.62 | ||||
| 5830443L24Rik | 0.60 | ||||
| Rlbp1l1 | 0.60 | ||||
| Lrp2 | 0.58 | ||||
| Spnb2 | 0.57 | ||||
| Sparc | 0.56 | ||||
| Smc1b | 0.54 | ||||
Differentially expressed genes were identified using limma package with a P-value threshold of 0.001. Categorization of genes (Groups A-G) corresponds to the same classification in Figure 1A. Values represent the fold change in linear scale.
The single gene differentially expressed to the greatest extent (9–12-fold increase) in all smoke-exposed groups as compared to controls was Cyp1b1 (Table 1), a Phase I detoxification enzyme involved in both the activation of carcinogens such as BaP and the metabolism of E2.
A surprising finding is the significant upregulation of Cryptochrome 1 (Cry1), one of the key transcriptional regulators of circadian rhythm, in response to smoke exposure. After Cyp1b1, Cry1 was the gene differentially regulated to the greatest extent following 3 wk of cigarette smoke exposure (fold-change = 1.99; Table 1) and one of the 10 genes altered at all 3 time points after smoke exposure (Fig. 1).
Quantitative Real-Time PCR and Western blot analysis
Differential expression of the genes that were modulated at all 3 time points and have a known function (7 of the 10 genes, Group A, Table 1) was validated by real-time PCR. All genes tested showed at least a 2-fold change in relative quantitation or had a FDR of less than 0.10, thereby validating the cDNA microarray results. The results for the 3, 8 and 20 wk time points are presented in Table 2. A strong correlation was observed between the fold-changes in gene expression determined by cDNA microarray and RT-PCR (Spearman's ρ was 0.9, 0.79 and 0.9 for samples obtained after 3, 8 and 20 wk of smoke exposure, respectively).
TABLE 2.
Gene expression analyses by quantitative real-time PCR
| 3 weeks | 8 weeks | 20 weeks | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Gene | P value | FDR | Fold-change | P value | FDR | Fold-change | P value | FDR | Fold-change |
| Cyp1b1 | 2.16E-03 | 4.33E-03 | 22.58 | 2.16E-03 | 2.89E-03 | 22.58 | 2.16E-03 | 3.46E-03 | 19.05 |
| Cry1 | 2.16E-03 | 4.33E-03 | 4.30 | 2.16E-03 | 2.89E-03 | 2.79 | 2.16E-03 | 3.46E-03 | 2.94 |
| Cbr3 | 2.16E-03 | 4.33E-03 | 3.56 | 2.16E-03 | 2.89E-03 | 2.55 | 2.16E-03 | 3.46E-03 | 2.79 |
| Ces3 | 6.49E-02 | 6.49E-02 | 2.38 | 8.66E-03 | 9.89E-03 | 1.68 | 4.11E-02 | 4.70E-02 | 1.57 |
| Tef | 2.16E-03 | 4.33E-03 | 2.53 | 6.49E-02 | 6.49E-02 | 1.54 | 4.33E-03 | 5.77E-03 | 1.87 |
| Ugt1a6a | 6.49E-02 | 6.49E-02 | 1.71 | 2.16E-03 | 2.89E-03 | 2.58 | 6.49E-02 | 6.49E-02 | 1.46 |
| Col3a1 | 1.52E-02 | 2.42E-02 | 0.54 | 2.16E-03 | 2.89E-03 | 0.30 | 2.16E-03 | 3.46E-03 | 0.38 |
| Hdc | 6.49E-02 | 6.49E-02 | 0.91 | 2.16E-03 | 2.89E-03 | 0.43 | 2.16E-03 | 3.46E-03 | 0.43 |
Total RNA was assayed by RT-PCR. Samples were analyzed in quadruplicate, and the resulting data were expressed as the average cycle threshold (Ct). The housekeeping gene Hprt1 was used for data normalization (ΔCt). Comparisons between control and smoke-exposed samples for statistical significance were determined using the Mann-Whitney test (P-value) and the step-up method of Benjamini & Hochberg (False Discovery Rate (FDR)). Fold-change was calculated using the Comparative Ct method (ΔΔCt). Genes induced or repressed following smoke exposure are indicated by a fold-change >1 or <1, respectively.
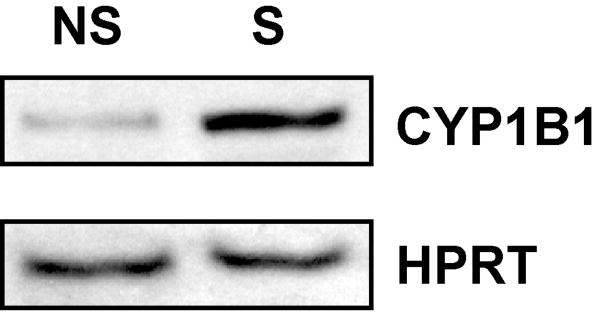
Examination of three commercial antibodies for their specificity for mouse Cyp1b1 indicated that none were highly specific for the protein of interest when analyzed by Western blot. For this reason, the ability of smoke to increase CYP1B1 mRNA expression was validated at the protein level using pooled preparations of human lung microsomes. Western blot analyses revealed a significant elevation in CYP1B1 protein in microsomes from smokers as compared to nonsmokers. These data not only confirm that both CYP1B1 mRNA and protein levels are elevated after smoke exposure but indicate the relevance of the murine finding to humans (Fig. 2).
Fig. 2. Western blot analysis of CYP1B1 in human pulmonary microsomes from nonsmokers (NS) and smokers (S).
Each sample (50 μg) contains a pool of microsomal protein from 4 individuals of mixed genders. HPRT was used as a loading control.
Identification of biological pathways and networks modulated by tobacco smoke
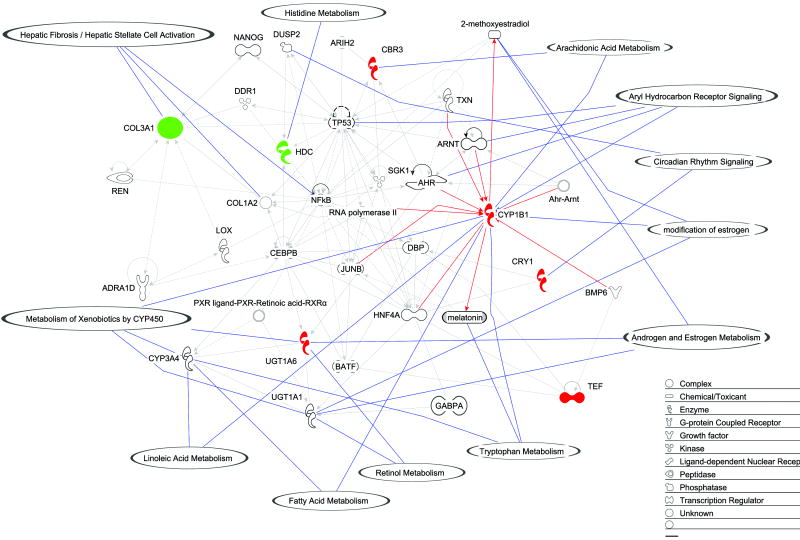
In addition to identifying individual genes differentially expressed (fold-change), a complementary strategy was employed to identify biological pathways and networks modulated by short-term exposure to tobacco smoke. The fold-changes of the 10 genes differentially expressed at all time points (3, 8 and 20 wk of smoke exposure) were mapped through a statistical computation method (Ingenuity Pathways Analysis Software). Fig. 3 depicts a high-scoring network based on 7 out of 10 genes that were eligible for network construction. In addition, canonical pathways and significant functions were mapped onto this network.
Fig. 3. Network of genes differentially expressed in common following 3, 8, and 20 wk of smoke exposure.
Differentially expressed genes (N=7) are depicted as a network with overlayed functions and pathways according to Ingenuity Pathways Analysis Software. The green and red colors represent down- and upregulated genes, respectively. The remaining genes are involved in the network through direct or indirect interactions. Red lines connecting Cyp1b1 to other genes indicate a direct relationship with Cyp1b1 (protein-protein interaction or protein-DNA). For example, the Ahr-Arnt complex increases transcription of Cyp1b1 in mammals (47). Solid blue lines with balloons indicate gene function or a pathway in which a gene is involved.
Consistent with induction of Cyp1b1 and its role in estrogen metabolism, oxidation of estrogen was identified as part of the network significantly modulated by tobacco smoke exposure, as highlighted in Fig. 3. Cyp1b1 was also present in several other pathways identified as being altered by smoke exposure including metabolism of xenobiotics by cytochrome P450, linoleic acid metabolism, fatty acid metabolism, and tryptophan metabolism.
Additional pathways modulated by smoke exposure are indicated in Fig. 3 and include hepatic fibrosis, histidine metabolism, arachidonic acid metabolism, AHR signaling and circadian rhythm signaling. Further studies are required to determine if either activation or repression of these biological pathways contributes to smoke-induced lung tumorigenesis.
Detection and localization of estrogens within murine lung tissue
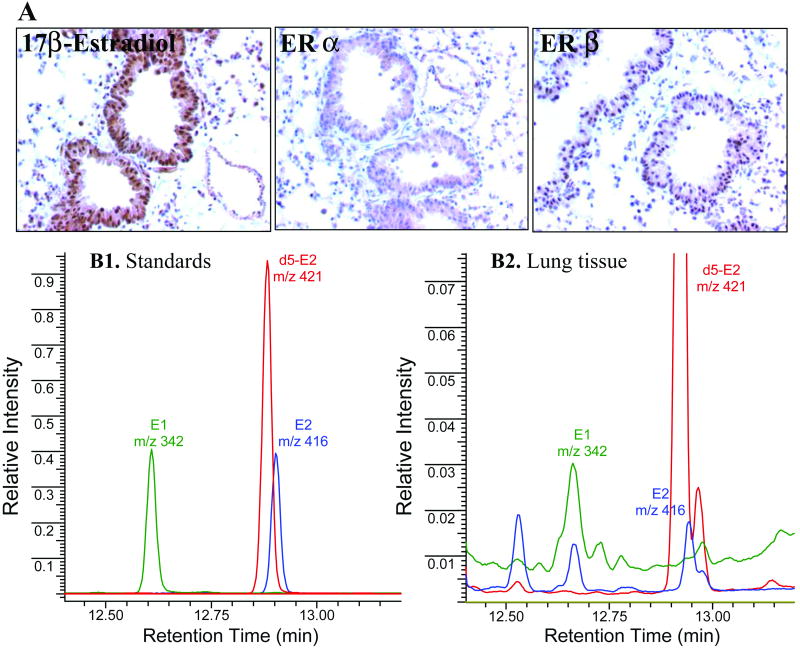
Although metabolism of estrogen is an important activity of Cyp1b1, this hormone had not been detected in murine lung tissue previously. We established a sensitive GC/MS method for the detection of E2 and E1, which can be converted to E2. Solvent extraction and GC/MS protocols were developed using standard solutions of the compounds E1 and E2. Lung tissue extracts were spiked with known amounts of E1 and E2 prior to extraction to assess recovery. Representative chromatograms of standards and extracts of lung tissue, illustrating the ions monitored, are presented in Fig. 4. Analysis of lung tissue from 8 untreated female mice clearly showed the presence of both E1 and E2 in extracts. The limit of detection per injection for E1 and E2 standards was 0.03 pmol, and the recoveries were 93% and 91%, respectively. However, the recovery of each compound from tissue was lower (E1 29% and E2 28%). Therefore, the actual concentration of estrogens in lung tissue could not be quantified due to low recovery. Efforts are underway to optimize the recovery and expand the methodology to measure a full panel of estrogen metabolites.
Fig. 4. Detection of estrogens within murine lung tissue.
Lung tissue from female A/J mice was subjected to immunohistochemical and GC/MS analyses. A. Detection of 17β-estradiol (E2), Erα and Erβ in lung epithelial cells by immunostaining. The bronchioloalveolar epithelium (BAE) stained positive for all antigens evaluated. Subcellular staining was observed as follows: E2, strong nuclear and cytoplasmic staining in the BAE and some pneumocytes; ERα, cytoplasmic staining of the BAE; and ERβ, nuclear staining in the BAE and some pneumocytes. B. Selective ion monitoring of trimethylsilyl derivatives of E1 and E2 (1.1 pmol each) and d5-E2 (2.6 pmol) as standards (B1) and in the murine lung tissue (B2). Each trace represents different ions monitored. Deuterium-labeled E2 represents the internal standard. Unmarked peaks in B2 denote unknowns; upper part of the chromatogram was cropped to enhance the visualization of small peaks.
The cellular localization of E2 within female murine lung tissue was determined by immunohistochemistry. Estrogen receptors (ERα and ERβ) were also examined to determine if ER-mediated estrogen signaling could occur within lung tissue. Staining for all antigens was localized primarily to the bronchial and bronchioloalveolar epithelium. Strong nuclear and cytoplasmic staining of E2 was observed, while staining of ERα and ERβ was localized primarily to the cytoplasm and nucleus, respectively (Fig. 4). No positive staining was detected in sections incubated with nonimmune IgG (negative control) (data not shown). Because this observation was purely qualitative, additional quantitative analyses of immunostained sections of untreated and smoke-exposed lung tissue are required to validate the observed subcellular localization of ER expression. Nevertheless, intracellular localization of E2 when combined with its detection by GC/MS in perfused lungs, as performed in this study, ensures that estrogens are present within murine lung tissue (as opposed to only in the circulation).
Discussion
Witschi and colleagues (15) have demonstrated that A/J mice that have been exposed to tobacco smoke for 20 wk develop an increased multiplicity of lung tumors when allowed to recover in ambient air for 16 wk prior to sacrifice as compared to those exposed to tobacco smoke continuously for 36 wk. This model suggests that early changes in gene expression within a smoke-exposed lung lead to irreversible cellular damage that is sufficient to initiate lung carcinogenesis. Thus, genes whose expression is altered very early in the carcinogenesis process represent cellular alterations that may serve as early targets for intervention in smoke-induced lung carcinogenesis. The expression of genes that were altered consistently after 3, 8 and 20 wk of active smoke exposure (Cyp1b1, Cry1, Cbr3, Ugt1a6a, Ces3, Tef, Hdc, Col3a1 (Fig. 1 Group A and Tables 1 and 2)), as determined by cDNA microarray, was validated by quantitative real-time PCR. It should be noted that not all of the genes with an established role in smoke-induced carcinogenesis were represented on the microarray, including Cyp1a1.
Cyp1b1, a Phase I enzyme, was the gene induced to the greatest extent within the lungs following continuous exposure to tobacco smoke. CYP1b1 transcript levels were elevated as early as 3 wk and remained upregulated for the duration of smoke exposure (20 wk) (Fig. 1, Tables 1 and 2). Upregulation of Cyp1b1 by tobacco smoke is of utmost interest because of its role in the metabolism of not only the PAHs found in tobacco smoke (e.g., BaP) but also E2, resulting in the generation of highly reactive catechols and quinone metabolites. These derivatives are known to form DNA adducts and cause genotoxicity (21, 22). Administration of 4-E2, the major product of CYP1B1 metabolism of estrogen, to Syrian hamsters and CD-1 mice has led to the development of renal cancer (23) and uterine adenocarcinomas (24), respectively. In humans, higher levels of 4-E2 are present in endometrial and breast cancers as compared to normal tissue (25).
Transcriptional induction of pulmonary Cyp1b1 by tobacco smoke has not been reported previously for A/J mice (14, 26). A 2-fold increase in Cyp1b1 mRNA levels was observed within the lungs of Sprague-Dawley rats exposed to mainstream tobacco smoke for 3 h, but surprisingly not after 3 wk of exposure (27). These preclinical data are consistent with reports of CYP1B1 induction in human smokers (28, 29). The dual role of Cyp1b1 in the metabolism of both constituents of tobacco smoke and estrogens, the pulmonary induction of Cyp1b1 by short-term tobacco smoke exposure, and validation of the response of CYP1B1 to tobacco smoke in human microsomes, as observed in the present study, provide strong support for the A/J mouse as a highly relevant model system in which to investigate the role of hormones in smoke-induced lung carcinogenesis.
Although results from previous studies suggest that ER-mediated signaling can occur within the lung, the present study is the first to report the detection of E1 and E2 within murine lung tissue. It is likely that tobacco smoke modulates estrogen levels within the lung by altering the expression of estrogen metabolism genes, including Cyp1b1. The GC/MS protocol established in this study focuses on the detection of parent estrogens. Based on the important role of estrogen signaling and estrogen metabolites in breast carcinogenesis, a detailed analysis of the biological significance of this hormone in lung tissue is warranted. Analysis of estrogen metabolites in human breast tissue by HPLC revealed that the levels of 4-E2 and 4-E1 as well as the quinone conjugates are significantly elevated in breast tissue from women with cancer as compared to women with benign disease (30). Although data on the detection of estrogens within tissue has been reported primarily for the breast, the ability of human bronchial epithelial BEAS-2B cells to metabolize estrogen has been demonstrated in vitro. Accumulation of 2- and 4-E2 was observed in lung cells treated with BaP in the presence of E2 (31). In summary, detection of hydroxylated metabolites and estrogen conjugates (methoxy and others) within the lungs as well as estrogen-associated DNA adducts is a promising tool to elucidate the mechanism by which estrogen induces lung carcinogenesis.
E2 as well as ERα and ERβ were detected immunohistochemically in murine bronchioloalveolar cells (Fig. 4). Atypical cytoplasmic localization of ERα was observed in the present study. Although this finding is in agreement with the reported cytoplasmic localization of ERα in human lung tissue (normal and tumor) (32) and normal human bronchial epithelial cells (13), the biological significance of extranuclear localization of ERα remains unclear. In breast cancer cells, the ER can be present in the cytoplasm or in the cell membrane where it binds to growth factor receptors, such as the EGF receptor, and exerts its signaling through downstream kinases (9). Although the functionality of ERs within the murine lung was not investigated in this study, numerous studies suggest that estrogen signaling within the lung can promote cell proliferation (13, 32, 33).
Data generated in the present study with the A/J mouse model complement the findings from several epidemiological and preclinical analyses that suggest the involvement of estrogen in lung carcinogenesis, either alone or in combination with smoking (7, 34, 35). Similar to humans, the effect of hormones on lung carcinogenesis in animals has been suggested (36, 37). The multiplicity of lung tumors in mice exposed to mainstream cigarette smoke early in life was significantly higher in females than in males (36). Moreover, exposure of mice to diethylstilbestrol, a synthetic estrogen, increases the incidence and multiplicity of lung tumors induced by urethane administration (37).
Following Cyp1b1, Cry1 was the gene most differentially expressed in the present study (Figs. 1 and 3, Table 1). Cry1 is one of the regulators of circadian rhythm, controlling physiological, biochemical, and behavioral functions with a periodicity of approximately 24 h. A prior study in rats exposed to tobacco smoke describes a distinct cyclic pattern of expression of other circadian rhythm genes such as Arntl, Dbp and Nr1d2 but not Cry1 (38). “Clock genes” are emerging as central players in cell cycle control and proliferation, and the altered expression of these genes has been observed in both endometrial (39) and breast (40) cancer. Interestingly, levels of melatonin, an important neuroendocrine output of circadian rhythm, are also affected by smoke exposure (41), and CYP1B1 is capable of metabolizing melatonin to 6-hydroxy melatonin (42). Our data depict the direct interaction between enzyme (Cyp1b1) and substrate (melatonin) and the overexpression of Cry1 (Fig. 3). These data, when combined, provide support for further evaluation of the role of the circadian rhythm genes in tobacco smoke-induced lung carcinogenesis.
Following Cyp1b1 and Cry1, the other genes differentially regulated at all time points following smoke exposure (Group A) are involved in metabolism of both endogenous and exogenous compounds (Cbr3, Ugt1a6a, Ces3, Hdc), signal transduction (Tef), and the extracellular matrix (Col3a), and their expression has been associated with other cancer types. Cbr3 (carbonyl reductase 3) catalyzes the reduction of many endogenous and xenobiotic carbonyl compounds, including steroids and prostaglandins, to their corresponding alcohols. Its expression is reduced in oral squamous cell carcinoma cells when compared to premalignant dysplasias and hyperplasias and has been associated with reduced cell growth and motility (43). Histidine decarboxylase (Hdc) is a member of the histidine metabolism pathway and is responsible for the biosynthesis of histamine. Hdc has been suggested as a new marker for neuroendocrine differentiation, inflammatory pathologies and several leukemias and highly malignant forms of cancer, including small cell lung carcinoma (44). The involvement of histamine in growth of mouse and rat tumors has been suggested (45). Thyrotrophic embryonic factor (tef) is a transcription factor that controls the expression of many enzymes and regulators involved in detoxification and drug metabolism, such as cytochrome P450 enzymes, carboxylesterases, and constitutive androstane receptor (46).
In summary, this study identifies gene expression changes that are induced by smoke exposure (3, 8 and 20 wk). It is the first to report the successful detection of estrogen within murine lung tissue and a network of Cyp1b1-associated genes that are modulated by smoke exposure. The ability of tobacco smoke to induce alterations in the expression of genes related to estrogen metabolism within the lung provides new insight into the molecular basis of smoke-induced lung cancer; in particular female lung cancer. Alteration of circadian rhythm and other pathways is also reported, which may represent novel targets for lung cancer prevention.
Acknowledgments
We wish to thank Radka Stoyanova, Ph.D. for her contribution to the data analyses, Ruth Holland for her assistance with the animal experiments, Bela Verma, M.S. for technical support, and the DNA Microarray Facility at Fox Chase Cancer Center for printing the arrays and providing advice. We thank Erica Golemis, Ph.D. for her critical review of this study. We also thank Maureen Climaldi for her excellent assistance in preparing this manuscript for publication.
Supported by grant CA-06927 from the National Cancer Institute, an appropriation from the Commonwealth of Pennsylvania, including a grant from the PA Department of Health, and the Jerome M. Spencer and Arnold Zaslow Family Foundation (Fox Chase Cancer Center); grants CA-96310, CA-118114 and CA-125152 from the National Cancer Institute, and the Agnes Brown Duggan Endowment Funds (University of Louisville); and the Conselho Nacional de Desenvolvimento Científico e Tecnológico, and Fundação de Amparo à Pesquisa do Estado de São Paulo (grants 98/14335-2 (CEPID) and 99/11962-9 and 99/07390-0) (Brazil).
Abbreviations
- E1
estrone
- E2
17β-estradiol
- 4-E2
4-hydroxyestradiol
- AHR
aryl hydrocarbon receptor
- BaP
benzo(a)pyrene
- Cry1
cryptochrome 1
- CYP1A1
cytochrome P4501A1
- CYP1B1
cytochrome P4501B1
- ER
estrogen receptor
- EROD
ethoxyresorufin deethylase
- FDR
False Discovery Rate
- GC/MS
gas chromatography coupled mass spectroscopy
- GSTM1
glutathione S-transferase M1
- HPRT
hypoxanthine-guanine phosphoribosyltransferase
- HRT
hormone replacement therapy
- LIMMA
Linear Models for Microarray Data
- LOWESS
Locally Weighted Regression and Smoothing Scatter
- NSCLC
non-small cell lung cancer
- PAH
polycyclic aromatic hydrocarbon
- TBST
Tris-buffered saline with Tween 20
Footnotes
The authors have no conflicts to disclose.
References
- 1.American Cancer Society. Cancer Facts and Figures. 2008. [Google Scholar]
- 2.Ries LAG, Eisner MP, Kosary CL, et al. SEER Cancer Statistics Review 1975-2001. National Cancer Institute; 2004. [Google Scholar]
- 3.Henschke CI, Yip R, Miettinen OS. Women's susceptibility to tobacco carcinogens and survival after diagnosis of lung cancer. JAMA. 2006;296:180–4. doi: 10.1001/jama.296.2.180. [DOI] [PubMed] [Google Scholar]
- 4.Belani CP, Marts S, Schiller J, Socinski MA. Women and lung cancer: epidemiology, tumor biology, and emerging trends in clinical research. Lung Cancer. 2007;55:15–23. doi: 10.1016/j.lungcan.2006.09.008. [DOI] [PubMed] [Google Scholar]
- 5.Stabile LP, Siegfried JM. Estrogen receptor pathways in lung cancer. Curr Oncol Rep. 2004;6:259–67. doi: 10.1007/s11912-004-0033-2. [DOI] [PubMed] [Google Scholar]
- 6.Liu Y, Inoue M, Sobue T, Tsugane S. Reproductive factors, hormone use and the risk of lung cancer among middle-aged never-smoking Japanese women: a large-scale population-based cohort study. Int J Cancer. 2005;117:662–6. doi: 10.1002/ijc.21229. [DOI] [PubMed] [Google Scholar]
- 7.Ganti AK, Sahmoun AE, Panwalkar AW, Tendulkar KK, Potti A. Hormone replacement therapy is associated with decreased survival in women with lung cancer. J Clin Oncol. 2006;24:59–63. doi: 10.1200/JCO.2005.02.9827. [DOI] [PubMed] [Google Scholar]
- 8.Raunio H, Hakkola J, Hukkanen J, et al. Expression of xenobiotic-metabolizing CYPs in human pulmonary tissue. Exp Toxicol Pathol. 1999;51:412–7. doi: 10.1016/S0940-2993(99)80031-1. [DOI] [PubMed] [Google Scholar]
- 9.Yager JD, Davidson NE. Estrogen carcinogenesis in breast cancer. N Engl J Med. 2006;354:270–82. doi: 10.1056/NEJMra050776. [DOI] [PubMed] [Google Scholar]
- 10.Barnea ER, MacLusky NJ, Naftolin F. Kinetics of catechol estrogen-estrogen receptor dissociation: a possible factor underlying differences in catechol estrogen biological activity. Steroids. 1983;41:643–56. doi: 10.1016/0039-128x(83)90030-2. [DOI] [PubMed] [Google Scholar]
- 11.Cavalieri E, Frenkel K, Liehr JG, Rogan E, Roy D. Estrogens as endogenous genotoxic agents--DNA adducts and mutations. J Natl Cancer Inst Monogr. 2000;27:75–93. doi: 10.1093/oxfordjournals.jncimonographs.a024247. [DOI] [PubMed] [Google Scholar]
- 12.Huber RM, Stratakis DF. Molecular oncology--perspectives in lung cancer. Lung Cancer. 2004;45(Suppl 2):S209–S2013. doi: 10.1016/j.lungcan.2004.07.973. [DOI] [PubMed] [Google Scholar]
- 13.Gordon T, Bosland M. Strain-dependent differences in susceptibility to lung cancer in inbred mice exposed to mainstream cigarette smoke. Cancer Lett. 2009;275:213–20. doi: 10.1016/j.canlet.2008.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Witschi H, Espiritu I, Dance ST, Miller MS. A mouse lung tumor model of tobacco smoke carcinogenesis. Toxicol Sci. 2002;68:322–30. doi: 10.1093/toxsci/68.2.322. [DOI] [PubMed] [Google Scholar]
- 15.Witschi H, Espiritu I, Peake JL, Wu K, Maronpot RR, Pinkerton KE. The carcinogenicity of environmental tobacco smoke. Carcinogenesis. 1997;18:575–86. doi: 10.1093/carcin/18.3.575. [DOI] [PubMed] [Google Scholar]
- 16.Lagueux J, Affar EB, Nadeau D, Ayotte P, Dewailly E, Poirier GG. A microassay for the detection of low levels of cytochrome P450 O-deethylation activities with alkoxyresorufin substrates. Mol Cell Biochem. 1997;175:125–9. doi: 10.1023/a:1006835712436. [DOI] [PubMed] [Google Scholar]
- 17.Gentleman RC, Carey VJ, Bates DM, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ritchie ME, Silver J, Oshlack A, et al. A comparison of background correction methods for two-colour microarrays. Bioinformatics. 2007;23:2700–7. doi: 10.1093/bioinformatics/btm412. [DOI] [PubMed] [Google Scholar]
- 19.Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004;3 doi: 10.2202/1544-6115.1027. Article 3. [DOI] [PubMed] [Google Scholar]
- 20.Benjamini Y, Hochberg Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. (B).J Royal Statistical Society. 1995;57:289–300. [Google Scholar]
- 21.Hecht SS. Tobacco carcinogens, their biomarkers and tobacco-induced cancer. Nat Rev Cancer. 2003;3:733–44. doi: 10.1038/nrc1190. [DOI] [PubMed] [Google Scholar]
- 22.Luch A. Nature and nurture - lessons from chemical carcinogenesis. Nat Rev Cancer. 2005;5:113–25. doi: 10.1038/nrc1546. [DOI] [PubMed] [Google Scholar]
- 23.Liehr JG, Fang WF, Sirbasku DA, Ari-Ulubelen A. Carcinogenicity of catechol estrogens in Syrian hamsters. J Steroid Biochem. 1986;24:353–6. doi: 10.1016/0022-4731(86)90080-4. [DOI] [PubMed] [Google Scholar]
- 24.Newbold RR, Liehr JG. Induction of uterine adenocarcinoma in CD-1 mice by catechol estrogens. Cancer Res. 2000;60:235–7. [PubMed] [Google Scholar]
- 25.Liehr JG, Ricci MJ. 4-Hydroxylation of estrogens as marker of human mammary tumors. Proc Natl Acad Sci USA. 1996;93:3294–6. doi: 10.1073/pnas.93.8.3294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Witschi H. A/J mouse as a model for lung tumorigenesis caused by tobacco smoke: strengths and weaknesses. Exp Lung Res. 2005;31:3–18. doi: 10.1080/01902140490494959. [DOI] [PubMed] [Google Scholar]
- 27.Gebel S, Gerstmayer B, Bosio A, Haussmann HJ, Van Miert E, Muller T. Gene expression profiling in respiratory tissues from rats exposed to mainstream cigarette smoke. Carcinogenesis. 2004;25:169–78. doi: 10.1093/carcin/bgg193. [DOI] [PubMed] [Google Scholar]
- 28.Spira A, Beane J, Shah V, et al. Effects of cigarette smoke on the human airway epithelial cell transcriptome. Proc Natl Acad Sci USA. 2004;101:10143–8. doi: 10.1073/pnas.0401422101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Port JL, Yamaguchi K, Du B, et al. Tobacco smoke induces CYP1B1 in the aerodigestive tract. Carcinogenesis. 2004;25:2275–81. doi: 10.1093/carcin/bgh243. [DOI] [PubMed] [Google Scholar]
- 30.Rogan EG, Badawi AF, Devanesan PD, et al. Relative imbalances in estrogen metabolism and conjugation in breast tissue of women with carcinoma: potential biomarkers of susceptibility to cancer. Carcinogenesis. 2003;24:697–702. doi: 10.1093/carcin/bgg004. [DOI] [PubMed] [Google Scholar]
- 31.Chang LW, Chang YC, Ho CC, Tsai MH, Lin P. Increase of carcinogenic risk via enhancement of cyclooxygenase-2 expression and hydroxyestradiol accumulation in human lung cells as a result of interaction between BaP and 17-beta estradiol. Carcinogenesis. 2007;28:1606–12. doi: 10.1093/carcin/bgm013. [DOI] [PubMed] [Google Scholar]
- 32.Stabile LP, Davis AL, Gubish CT, et al. Human non-small cell lung tumors and cells derived from normal lung express both estrogen receptor α and β and show biological responses to estrogen. Cancer Res. 2002;62:2141–50. [PubMed] [Google Scholar]
- 33.Marquez-Garban DC, Chen HW, Fishbein MC, Goodglick L, Pietras RJ. Estrogen receptor signaling pathways in human non-small cell lung cancer. Steroids. 2007;72:135–43. doi: 10.1016/j.steroids.2006.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chlebowski RT, Schwartz A, Wakelee H, et al. Non-small cell lung cancer and estrogen plus progestin use in postmenopausal women in the Women's Health Initiative randomized clinical trial. J Clin Oncol. 2009;27(18S) Abstract CRA1500. [Google Scholar]
- 35.Taioli E, Wynder EL. Endocrine factors and adenocarcinoma of the lung in women. J Natl Cancer Inst. 1994;86:869–70. doi: 10.1093/jnci/86.11.869. Letter. [DOI] [PubMed] [Google Scholar]
- 36.Balansky R, Ganchev G, Iltcheva M, Steele VE, D'Agostini F, De Flora S. Potent carcinogenicity of cigarette smoke in mice exposed early in life. Carcinogenesis. 2007;28:2236–43. doi: 10.1093/carcin/bgm122. [DOI] [PubMed] [Google Scholar]
- 37.Jiang YG, Chen JK, Wu ZL. Promotive effect of diethylstilbestrol on urethan-induced mouse lung tumorigenesis. Chemosphere. 2000;41:187–90. doi: 10.1016/s0045-6535(99)00410-5. [DOI] [PubMed] [Google Scholar]
- 38.Gebel S, Gerstmayer B, Kuhl P, Borlak J, Meurrens K, Muller T. The kinetics of transcriptomic changes induced by cigarette smoke in rat lungs reveals a specific program of defense, inflammation, and circadian clock gene expression. Toxicol Sci. 2006;93:422–31. doi: 10.1093/toxsci/kfl071. [DOI] [PubMed] [Google Scholar]
- 39.Yeh KT, Yang MY, Liu TC, et al. Abnormal expression of period 1 (PER1) in endometrial carcinoma. J Pathol. 2005;206:111–20. doi: 10.1002/path.1756. [DOI] [PubMed] [Google Scholar]
- 40.Chen ST, Choo KB, Hou MF, Yeh KT, Kuo SJ, Chang JG. Deregulated expression of the PER1, PER2 and PER3 genes in breast cancers. Carcinogenesis. 2005;26:1241–6. doi: 10.1093/carcin/bgi075. [DOI] [PubMed] [Google Scholar]
- 41.Ozguner F, Koyu A, Cesur G. Active smoking causes oxidative stress and decreases blood melatonin levels. Toxicol Ind Health. 2005;21:21–6. doi: 10.1191/0748233705th211oa. [DOI] [PubMed] [Google Scholar]
- 42.Ma X, Idle JR, Krausz KW, Gonzalez FJ. Metabolism of melatonin by human cytochromes p450. Drug Metab Dispos. 2005;33:489–94. doi: 10.1124/dmd.104.002410. [DOI] [PubMed] [Google Scholar]
- 43.Ohkura-Hada S, Kondoh N, Hada A, et al. Carbonyl reductase 3 (CBR3) mediates 9-cis-retinoic acid-induced cytostatis and is a potential prognostic marker for oral malignancy. Open Dent J. 2008;2:78–88. doi: 10.2174/1874210600802010078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Graff L, Frungieri M, Zanner R, Pohlinger A, Prinz C, Gratzl M. Expression of histidine decarboxylase and synthesis of histamine by human small cell lung carcinoma. Am J Pathol. 2002;160:1561–5. doi: 10.1016/S0002-9440(10)61102-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bartholeyns J, Bouclier M. Involvement of histamine in growth of mouse and rat tumors: antitumoral properties of monofluoromethylhistidine, an enzyme-activated irreversible inhibitor of histidine decarboxylase. Cancer Res. 1984;44:639–45. [PubMed] [Google Scholar]
- 46.Gachon F, Olela FF, Schaad O, Descombes P, Schibler U. The circadian PAR-domain basic leucine zipper transcription factors DBP, TEF, and HLF modulate basal and inducible xenobiotic detoxification. Cell Metab. 2006;4:25–36. doi: 10.1016/j.cmet.2006.04.015. [DOI] [PubMed] [Google Scholar]
- 47.Matthews J, Gustafsson JA. Estrogen receptor and aryl hydrocarbon receptor signaling pathways. Nucl Recept Signal. 2006;4:e016. doi: 10.1621/nrs.04016. [DOI] [PMC free article] [PubMed] [Google Scholar]